### connect the drive
from google.colab import drive
drive.mount("/content/drive/")
Drive already mounted at /content/drive/; to attempt to forcibly remount, call drive.mount("/content/drive/", force_remount=True).
### set working directory....
%cd "/content/drive/MyDrive/Yemaachi_works"
/content/drive/MyDrive/Yemaachi_works
### import biopython:
!pip install biopython
### install gspread:
!pip install --upgrade -q gspread
Collecting biopython
Downloading biopython-1.79-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.whl (2.3 MB)
�[K |████████████████████████████████| 2.3 MB 5.0 MB/s
�[?25hRequirement already satisfied: numpy in /usr/local/lib/python3.7/dist-packages (from biopython) (1.21.5)
Installing collected packages: biopython
Successfully installed biopython-1.79
#### rating the limit of data reads
!pip install -q ratelimit
Building wheel for ratelimit (setup.py) ... �[?25l�[?25hdone
##### check for the lists of folders in the directory:
#%ls -sh "/content/drive/MyDrive/Yemaachi_works/Cyclophosphamide /"
%ls -sh "/content/drive/MyDrive/Yemaachi_works/Cyclophosphamide "
total 932K
3.0K all_CategoriesGenes.csv
512 all_CategoriesGenes.gsheet
15K all-data_cyclophosphamide_clinical.tsv
182K all-data_cyclophosphamide_variants.tsv
512 all_data_snp.gsheet
15K all-data.tsv
235K all_interactions_dataframe.csv
512 all_interactions_dataframe.gsheet
4.0K �[0m�[01;34mclinical_annotations_allele�[0m/
4.0K clinical_annotations.csv
44K cpic_drug_recommendation.csv
512 cpic_drug_recommendation.gsheet
512 CYCLOPHOSPHAMIDE_genes.gsheet
21K CYCLOPHOSPHAMIDE.xlsx
512 'known_genes_cyclophosphamide (1).gsheet'
8.0K known_genes_cyclophosphamide.csv
512 known_genes_cyclophosphamide.gsheet
1.0K 'PA2034 (1).tsv'
75K PA2034.pdf
108K PA2035.pdf
3.0K PA2035.tsv
4.0K �[01;34mupdate_allele_data�[0m/
512 'variants_annotations (1).gsheet'
37K variants_annotations.csv
512 variants_annotations.gsheet
512 'variants_genes_cyclophosphamide (1).gsheet'
512 'variants_genes_cyclophosphamide (2).gsheet'
170K variants_genes_cyclophosphamide.csv
512 variants_genes_cyclophosphamide.gsheet
Pipeline creation:
- Developing of pipeline for data extraction.
### import the modules and packages needed
import pandas as pd
import numpy as np
from glob import glob
import requests
import re
import json
from urllib.request import urlretrieve
import Bio
from Bio import SeqIO, SearchIO, Entrez
from Bio.Seq import Seq
from Bio.SeqUtils import GC
from Bio.Blast import NCBIWWW
from Bio.Data import CodonTable
from ratelimit import limits
import time
from typing import List, Any
#### check the version of biopython
print(Bio.__version__)
1.79
### import already scraped data info for gene metabolism
capecitabine_genes = pd.read_csv("/content/drive/MyDrive/Yemaachi_works/Capecitabine/CAPECITABINE_genes.csv")
capecitabine_genes.head()
|
Unnamed: 0 |
interactionId |
interactionTypes |
geneName |
geneLongName |
geneEntrezId |
sources |
pmids |
score |
| 0 |
0 |
8fdee0e9-a54e-4d52-9be1-59e18866e3c6 |
['inhibitor'] |
TYMS |
THYMIDYLATE SYNTHETASE |
7298 |
['ClearityFoundationBiomarkers', 'ClearityFoun... |
[15134221, 16926630, 15866500, 15132128, 11752... |
0.48 |
| 1 |
1 |
ba94c9df-0282-42cb-99bd-1ef4729443c0 |
[] |
ERCC1 |
ERCC EXCISION REPAIR 1, ENDONUCLEASE NON-CATAL... |
2067 |
['PharmGKB'] |
[25026457] |
0.21 |
| 2 |
2 |
2b3a2aa9-54a7-4f8e-ba74-bc6fb919d61b |
[] |
MGAT4A |
MANNOSYL (ALPHA-1,3-)-GLYCOPROTEIN BETA-1,4-N-... |
11320 |
['PharmGKB'] |
[26222057] |
0.89 |
| 3 |
3 |
f9d41d4d-c37c-4e95-b991-5d839a2f2402 |
[] |
DLG5 |
DISCS LARGE MAGUK SCAFFOLD PROTEIN 5 |
9231 |
['PharmGKB'] |
[] |
0.89 |
| 4 |
4 |
73600292-d86a-4047-b1e0-16bac81b5b88 |
[] |
ABCG2 |
ATP BINDING CASSETTE SUBFAMILY G MEMBER 2 (JUN... |
9429 |
['PharmGKB'] |
[24338217, 20530282] |
0.09 |
capecitabine_genes.columns
Index(['Unnamed: 0', 'interactionId', 'interactionTypes', 'geneName',
'geneLongName', 'geneEntrezId', 'sources', 'pmids', 'score'],
dtype='object')
capecitabine_genes.shape
(36, 9)
capecitabine_genes.geneName.value_counts()
TYMS 1
MTHFR 1
KRAS 1
CES1P1 1
HLA-G 1
ABCG2 1
CDH1 1
TP53 1
CES1 1
SELE 1
PIK3CA 1
BRAF 1
CDA 1
UMPS 1
PTGS2 1
ERBB2 1
CYP1A1 1
DLG5 1
MET 1
VEGFA 1
DPYD 1
ENOSF1 1
MGAT4A 1
AREG 1
ERCC1 1
ADCY2 1
MIR2054 1
REV3L 1
CYP19A1 1
SLC22A7 1
ABCB1 1
EXO1 1
MIR27A 1
PTEN 1
TYMP 1
TYMSOS 1
Name: geneName, dtype: int64
### test pmids
capecitabine_genes.pmids[4][1:-1].split(",")
['24338217', ' 20530282']
capecitabine_genes.pmids[0][1:-1]
'15134221, 16926630, 15866500, 15132128, 11752352, 15709193'
#### extract all pmids
pmids_ = []
genes = []
for x,y in zip(capecitabine_genes.geneName,capecitabine_genes.pmids):
if len(y[1:-1]) == 0:
continue
else:
new_pmids = y[1:-1].split(",")
for pmid in new_pmids:
pmids_.append(pmid.strip())
genes.append(x)
#### combine feature together
genes_pmids = pd.DataFrame()
genes_pmids["genes"] = genes
genes_pmids["pmids"] = pmids_
genes_pmids.head()
|
genes |
pmids |
| 0 |
TYMS |
15134221 |
| 1 |
TYMS |
16926630 |
| 2 |
TYMS |
15866500 |
| 3 |
TYMS |
15132128 |
| 4 |
TYMS |
11752352 |
capecitabine_genes.sources[0][1:-1]
"'ClearityFoundationBiomarkers', 'ClearityFoundationClinicalTrial', 'ChemblInteractions', 'GuideToPharmacology'"
genes_pmids.shape
(98, 2)
urlink = requests.get("https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/15134221/unicode").json()
urlink
{'date': '20190911',
'documents': [{'id': '15134221',
'infons': {},
'passages': [{'annotations': [],
'infons': {'type': 'title'},
'offset': 0,
'relations': [],
'sentences': [],
'text': 'Potential for predicting toxicity and response of fluoropyrimidines in patients.'},
{'annotations': [],
'infons': {'type': 'abstract'},
'offset': 81,
'relations': [],
'sentences': [],
'text': 'The efficacy of cancer therapy is compromised by the fact that there are currently no good ways to predict which patients will benefit from treatment. This long standing goal is closer to becoming a reality as more is learned about the molecules that affect the activities of various therapeutic agents. The fluoropyrimidine antimetabolites drugs have been in clinical use for over 4 decades and the cellular proteins important for their activities have been studied in detail. The most important are the major target enzyme, thymidylate synthase (TS) and the rate limiting enzyme in the degradation pathway, dihydropyrimidine dehydrogenase (DPD), equally important for the analogue capecitabine is thymidine phosphorylase (TP), which is rate limiting for activation of this prodrug. A number of assays are available for these enzymes, including enzyme activity measurements. quantitative PCR for RNA expression and immunological methods for protein expression. With each of these methods, more clinical studies are required to validate their clinical usefulness.'}],
'relations': []}],
'infons': {},
'key': 'collection.key',
'source': 'PubMed'}
urlink["documents"][0]["passages"][1]["text"]
'The efficacy of cancer therapy is compromised by the fact that there are currently no good ways to predict which patients will benefit from treatment. This long standing goal is closer to becoming a reality as more is learned about the molecules that affect the activities of various therapeutic agents. The fluoropyrimidine antimetabolites drugs have been in clinical use for over 4 decades and the cellular proteins important for their activities have been studied in detail. The most important are the major target enzyme, thymidylate synthase (TS) and the rate limiting enzyme in the degradation pathway, dihydropyrimidine dehydrogenase (DPD), equally important for the analogue capecitabine is thymidine phosphorylase (TP), which is rate limiting for activation of this prodrug. A number of assays are available for these enzymes, including enzyme activity measurements. quantitative PCR for RNA expression and immunological methods for protein expression. With each of these methods, more clinical studies are required to validate their clinical usefulness.'
from tqdm import tqdm_notebook
from tqdm.notebook import tqdm
from time import sleep
#### retrieve file
def retrieveDocuments(pmids=None):
"""
Args:
Takes the lists of all pmids for documents given the genes.
"""
abstract_doc = [] ##### initial empy list to access abstract text
title_doc = [] ##### the title of the article.
url = "https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/"
unicode = "/unicode"
test = 0
for pmid in tqdm(pmids,desc="collections of files:"):
urlink = url + str(pmid) + unicode
test += 1
print(f"link {test}: {urlink}") ### print the links
### make a query to through the api
jsonfile = requests.get(urlink).json()
if len(jsonfile["documents"][0]["passages"]) == 1:
### check the num of elements in the list....
title = jsonfile["documents"][0]["passages"][0]["text"]
text = "No abstract"
title_doc.append(title)
abstract_doc.append(text)
else:
### extract the abstract the title and abstract
text = jsonfile["documents"][0]["passages"][1]["text"]
title = jsonfile["documents"][0]["passages"][0]["text"]
abstract_doc.append(text)
title_doc.append(title)
sleep(0.1)
return abstract_doc,title_doc
### access the files.....
doc_abstract , title_doc = retrieveDocuments(genes_pmids.pmids)
collections of files:: 0%| | 0/78 [00:00<?, ?it/s]
link 1: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/15134221/unicode
link 2: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/16926630/unicode
link 3: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/15866500/unicode
link 4: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/15132128/unicode
link 5: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/11752352/unicode
link 6: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/15709193/unicode
link 7: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/25026457/unicode
link 8: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/26222057/unicode
link 9: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/24338217/unicode
link 10: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/20530282/unicode
link 11: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/26487584/unicode
link 12: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/24980946/unicode
link 13: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/17549067/unicode
link 14: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/27995989/unicode
link 15: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/20714149/unicode
link 16: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/27001118/unicode
link 17: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/23263912/unicode
link 18: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/28347776/unicode
link 19: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/28139840/unicode
link 20: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/16279094/unicode
link 21: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/23988873/unicode
link 22: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/22426923/unicode
link 23: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/28139840/unicode
link 24: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/24957073/unicode
link 25: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/27738344/unicode
link 26: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/18245544/unicode
link 27: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/17700593/unicode
link 28: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/24167597/unicode
link 29: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/29845393/unicode
link 30: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/27864592/unicode
link 31: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/18299612/unicode
link 32: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/20819423/unicode
link 33: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/27995989/unicode
link 34: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/19384296/unicode
link 35: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/29134491/unicode
link 36: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/23314736/unicode
link 37: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/20638924/unicode
link 38: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/25331073/unicode
link 39: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/20385995/unicode
link 40: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/27557140/unicode
link 41: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/26967565/unicode
link 42: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/23407049/unicode
link 43: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/26014925/unicode
link 44: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/23736036/unicode
link 45: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/20647221/unicode
link 46: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/28347776/unicode
link 47: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/16818689/unicode
link 48: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/25026457/unicode
link 49: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/19219602/unicode
link 50: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/26920887/unicode
link 51: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/26432108/unicode
link 52: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/24167597/unicode
link 53: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/20125120/unicode
link 54: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/25955730/unicode
link 55: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/21325291/unicode
link 56: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/24167597/unicode
link 57: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/28347776/unicode
link 58: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/18473752/unicode
link 59: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/23736036/unicode
link 60: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/20504363/unicode
link 61: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/17192538/unicode
link 62: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/17679724/unicode
link 63: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/19289619/unicode
link 64: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/25287822/unicode
link 65: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/26432108/unicode
link 66: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/26487584/unicode
link 67: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/26920887/unicode
link 68: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/25677447/unicode
link 69: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/26804235/unicode
link 70: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/25782327/unicode
link 71: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/24401318/unicode
link 72: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/25655103/unicode
link 73: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/28347776/unicode
link 74: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/19571295/unicode
link 75: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/22180495/unicode
link 76: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/21142915/unicode
link 77: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/22026922/unicode
link 78: https://www.ncbi.nlm.nih.gov/research/bionlp/RESTful/pubmed.cgi/BioC_json/25815774/unicode
print(f"Total number extracted: \n \t\t {len(doc_abstract)} abstracts \n \t \t {len(title_doc)} titles")
Total number extracted:
78 abstracts
78 titles
genes_pmids["doc_abstract"] = doc_abstract
genes_pmids["title_doc"] = title_doc
genes_pmids.head()
|
genes |
pmids |
doc_abstract |
title_doc |
| 0 |
TYMS |
15134221 |
The efficacy of cancer therapy is compromised ... |
Potential for predicting toxicity and response... |
| 1 |
TYMS |
16926630 |
The current reference treatment of hormone-ref... |
Synergistic cytotoxic interaction in hormone-r... |
| 2 |
TYMS |
15866500 |
A novel method employing high-performance liqu... |
Rapid quantitation of plasma 2'-deoxyuridine b... |
| 3 |
TYMS |
15132128 |
PURPOSE: The fluoropyrimidine carbamate (capec... |
Enzyme expression profiles suggest the novel t... |
| 4 |
TYMS |
11752352 |
A number of proteins and nucleic acids have be... |
TTD: Therapeutic Target Database. |
#### save the file
genes_pmids.to_csv("/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_genes_paper_summary.csv",index=False)
%pwd
'/content/drive/My Drive/Yemaachi_works'
!pip install metapub
Collecting metapub
�[?25l Downloading https://files.pythonhosted.org/packages/50/b7/ac81339f463d123fdd5131c3813d7e5a9b4f2c902c18e93974bd4c42e7f8/metapub-0.5.5.tar.gz (120kB)
�[K |████████████████████████████████| 122kB 6.7MB/s
�[?25hRequirement already satisfied: setuptools in /usr/local/lib/python3.7/dist-packages (from metapub) (57.0.0)
Requirement already satisfied: lxml in /usr/local/lib/python3.7/dist-packages (from metapub) (4.2.6)
Requirement already satisfied: requests in /usr/local/lib/python3.7/dist-packages (from metapub) (2.23.0)
Collecting eutils
�[?25l Downloading https://files.pythonhosted.org/packages/ae/05/292de2bc244d0f5cc900bd9d63d9c3cf16dd57684859873f1c6eba4771b1/eutils-0.6.0-py2.py3-none-any.whl (41kB)
�[K |████████████████████████████████| 51kB 5.9MB/s
�[?25hCollecting habanero
Downloading https://files.pythonhosted.org/packages/23/d5/5b3ecf668b50839028fbeb5c551a58af31c13e5a08bba0b19194670a4d16/habanero-0.7.4-py2.py3-none-any.whl
Requirement already satisfied: tabulate in /usr/local/lib/python3.7/dist-packages (from metapub) (0.8.9)
Collecting cssselect
Downloading https://files.pythonhosted.org/packages/3b/d4/3b5c17f00cce85b9a1e6f91096e1cc8e8ede2e1be8e96b87ce1ed09e92c5/cssselect-1.1.0-py2.py3-none-any.whl
Collecting unidecode
�[?25l Downloading https://files.pythonhosted.org/packages/9e/25/723487ca2a52ebcee88a34d7d1f5a4b80b793f179ee0f62d5371938dfa01/Unidecode-1.2.0-py2.py3-none-any.whl (241kB)
�[K |████████████████████████████████| 245kB 9.7MB/s
�[?25hRequirement already satisfied: docopt in /usr/local/lib/python3.7/dist-packages (from metapub) (0.6.2)
Requirement already satisfied: six in /usr/local/lib/python3.7/dist-packages (from metapub) (1.15.0)
Collecting tox
�[?25l Downloading https://files.pythonhosted.org/packages/d8/63/2fa635ac1b8a22e960654b07c270dfb53eb873aba261006536de40327b18/tox-3.23.1-py2.py3-none-any.whl (85kB)
�[K |████████████████████████████████| 92kB 6.8MB/s
�[?25hRequirement already satisfied: pytest in /usr/local/lib/python3.7/dist-packages (from metapub) (3.6.4)
Collecting coloredlogs
�[?25l Downloading https://files.pythonhosted.org/packages/a7/06/3d6badcf13db419e25b07041d9c7b4a2c331d3f4e7134445ec5df57714cd/coloredlogs-15.0.1-py2.py3-none-any.whl (46kB)
�[K |████████████████████████████████| 51kB 5.5MB/s
�[?25hCollecting python-Levenshtein
�[?25l Downloading https://files.pythonhosted.org/packages/2a/dc/97f2b63ef0fa1fd78dcb7195aca577804f6b2b51e712516cc0e902a9a201/python-Levenshtein-0.12.2.tar.gz (50kB)
�[K |████████████████████████████████| 51kB 5.4MB/s
�[?25hRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests->metapub) (3.0.4)
Requirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests->metapub) (2021.5.30)
Requirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests->metapub) (1.24.3)
Requirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests->metapub) (2.10)
Requirement already satisfied: pytz in /usr/local/lib/python3.7/dist-packages (from eutils->metapub) (2018.9)
Requirement already satisfied: tqdm in /usr/local/lib/python3.7/dist-packages (from habanero->metapub) (4.41.1)
Collecting pluggy>=0.12.0
Downloading https://files.pythonhosted.org/packages/a0/28/85c7aa31b80d150b772fbe4a229487bc6644da9ccb7e427dd8cc60cb8a62/pluggy-0.13.1-py2.py3-none-any.whl
Collecting virtualenv!=20.0.0,!=20.0.1,!=20.0.2,!=20.0.3,!=20.0.4,!=20.0.5,!=20.0.6,!=20.0.7,>=16.0.0
�[?25l Downloading https://files.pythonhosted.org/packages/03/08/f819421002e85a71d58368f7bffbe0b1921325e0e8ca7857cb5fb0e1f7c1/virtualenv-20.4.7-py2.py3-none-any.whl (7.2MB)
�[K |████████████████████████████████| 7.2MB 11.1MB/s
�[?25hRequirement already satisfied: packaging>=14 in /usr/local/lib/python3.7/dist-packages (from tox->metapub) (20.9)
Requirement already satisfied: toml>=0.9.4 in /usr/local/lib/python3.7/dist-packages (from tox->metapub) (0.10.2)
Requirement already satisfied: py>=1.4.17 in /usr/local/lib/python3.7/dist-packages (from tox->metapub) (1.10.0)
Requirement already satisfied: filelock>=3.0.0 in /usr/local/lib/python3.7/dist-packages (from tox->metapub) (3.0.12)
Requirement already satisfied: importlib-metadata>=0.12; python_version < "3.8" in /usr/local/lib/python3.7/dist-packages (from tox->metapub) (4.5.0)
Requirement already satisfied: attrs>=17.4.0 in /usr/local/lib/python3.7/dist-packages (from pytest->metapub) (21.2.0)
Requirement already satisfied: atomicwrites>=1.0 in /usr/local/lib/python3.7/dist-packages (from pytest->metapub) (1.4.0)
Requirement already satisfied: more-itertools>=4.0.0 in /usr/local/lib/python3.7/dist-packages (from pytest->metapub) (8.8.0)
Collecting humanfriendly>=9.1
�[?25l Downloading https://files.pythonhosted.org/packages/92/7e/a06472f484fa589933f39bfb41a7b849ca49f6d8e4fdfe978e27f0e3075c/humanfriendly-9.2-py2.py3-none-any.whl (86kB)
�[K |████████████████████████████████| 92kB 10.3MB/s
�[?25hRequirement already satisfied: appdirs<2,>=1.4.3 in /usr/local/lib/python3.7/dist-packages (from virtualenv!=20.0.0,!=20.0.1,!=20.0.2,!=20.0.3,!=20.0.4,!=20.0.5,!=20.0.6,!=20.0.7,>=16.0.0->tox->metapub) (1.4.4)
Collecting distlib<1,>=0.3.1
�[?25l Downloading https://files.pythonhosted.org/packages/87/26/f6a23dd3e578132cf924e0dd5d4e055af0cd4ab43e2a9f10b7568bfb39d9/distlib-0.3.2-py2.py3-none-any.whl (338kB)
�[K |████████████████████████████████| 348kB 41.3MB/s
�[?25hRequirement already satisfied: pyparsing>=2.0.2 in /usr/local/lib/python3.7/dist-packages (from packaging>=14->tox->metapub) (2.4.7)
Requirement already satisfied: zipp>=0.5 in /usr/local/lib/python3.7/dist-packages (from importlib-metadata>=0.12; python_version < "3.8"->tox->metapub) (3.4.1)
Requirement already satisfied: typing-extensions>=3.6.4; python_version < "3.8" in /usr/local/lib/python3.7/dist-packages (from importlib-metadata>=0.12; python_version < "3.8"->tox->metapub) (3.7.4.3)
Building wheels for collected packages: metapub, python-Levenshtein
Building wheel for metapub (setup.py) ... �[?25l�[?25hdone
Created wheel for metapub: filename=metapub-0.5.5-cp37-none-any.whl size=135372 sha256=644c3ecde753d9d2c77983a11c8b6438f261842d93236dcb9511f8672c452725
Stored in directory: /root/.cache/pip/wheels/af/d9/fc/c2f13edf0a7d0d335a69417183acfb3e08896743a79067cbf3
Building wheel for python-Levenshtein (setup.py) ... �[?25l�[?25hdone
Created wheel for python-Levenshtein: filename=python_Levenshtein-0.12.2-cp37-cp37m-linux_x86_64.whl size=149801 sha256=7487382130c4e69ca46cec3513536da97ea399becc3755218764707ed180df45
Stored in directory: /root/.cache/pip/wheels/b3/26/73/4b48503bac73f01cf18e52cd250947049a7f339e940c5df8fc
Successfully built metapub python-Levenshtein
�[31mERROR: pytest 3.6.4 has requirement pluggy<0.8,>=0.5, but you'll have pluggy 0.13.1 which is incompatible.�[0m
�[31mERROR: datascience 0.10.6 has requirement folium==0.2.1, but you'll have folium 0.8.3 which is incompatible.�[0m
Installing collected packages: eutils, habanero, cssselect, unidecode, pluggy, distlib, virtualenv, tox, humanfriendly, coloredlogs, python-Levenshtein, metapub
Found existing installation: pluggy 0.7.1
Uninstalling pluggy-0.7.1:
Successfully uninstalled pluggy-0.7.1
Successfully installed coloredlogs-15.0.1 cssselect-1.1.0 distlib-0.3.2 eutils-0.6.0 habanero-0.7.4 humanfriendly-9.2 metapub-0.5.5 pluggy-0.13.1 python-Levenshtein-0.12.2 tox-3.23.1 unidecode-1.2.0 virtualenv-20.4.7
from metapub import FindIt,PubMedFetcher,MedGenFetcher,MedGenConcept
fetch = PubMedFetcher()
pmids = ["29938344","15746054","17638512","8242617",
"20179710","21821736","24533712","20179710",
"27785604","29938344","25589624","29938344",
"27234217","25008867","20568049","19696793","29938344",
"10469894","16822847","17388661"]
for pmid in pmids:
article = fetch.article_by_pmid(pmid)
print(article.abstract)
print(article.journal)
print(article.doi)
print(article.chemicals)
print(article.url)
print()
PURPOSE: Cyclophosphamide and doxorubicin (adjuvant chemotherapy) are commonly used to treat breast cancer patients. Variation in the genes involved in pharmacodynamics and pharmacokinetics of these drugs plays an important role in prediction of drug response and survival. The present study was carried out with an aim to evaluate the variation in all the genes involved in pharmacokinetic and pharmacodynamics pathways of cyclophosphamide and doxorubicin, and correlate specific variants with disease outcome in breast cancer patients from the Malwa region of Punjab.
METHODS: A total of 250 confirmed breast cancer patients were involved in the study. Genotyping was performed on an Illumina Infinium HD assay platform using a Global Screening Array (GSA) microchip. GenomeStudio (Illumina, Inc.) was used for data preprocessing and a p value less than or equal to 5 × 10-8 was considered statistically significant. To rule out the influence of confounding risk factors, a step-wise multivariate regression analysis was carried out to evaluate the association of genotype with overall clinical outcome.
RESULTS: Two gene variants, CYP2C19 (G681A) and ALDH1A1*2 (17 bp deletion), were found to be significantly associated with the disease outcome, including overall survival, recurrence and metastasis, in breast cancer patients on adjuvant therapy. Both these genes are involved in the pharmacokinetics of cyclophosphamide. However, none of the variants in the genes involved in pharmacokinetics and pharmacodynamics of doxorubicin were found to have any significant impact on disease outcome in the studied group.
CONCLUSION: CYP2C19 (G681A) variant and ALDH1A1*2 emerged as two important biomarkers associated with bad outcome in breast cancer patients on adjuvant therapy.
Eur J Clin Pharmacol
10.1007/s00228-018-2505-6
{'D000903': {'substance_name': 'Antibiotics, Antineoplastic', 'registry_number': '0'}, 'D018906': {'substance_name': 'Antineoplastic Agents, Alkylating', 'registry_number': '0'}, 'D014408': {'substance_name': 'Biomarkers, Tumor', 'registry_number': '0'}, 'D004317': {'substance_name': 'Doxorubicin', 'registry_number': '80168379AG'}, 'D003520': {'substance_name': 'Cyclophosphamide', 'registry_number': '8N3DW7272P'}, 'C045793': {'substance_name': 'CYP2C19 protein, human', 'registry_number': 'EC 1.14.14.1'}, 'D065731': {'substance_name': 'Cytochrome P-450 CYP2C19', 'registry_number': 'EC 1.14.14.1'}, 'D000080924': {'substance_name': 'Aldehyde Dehydrogenase 1 Family', 'registry_number': 'EC 1.2.1'}, 'D000444': {'substance_name': 'Aldehyde Dehydrogenase', 'registry_number': 'EC 1.2.1.3'}, 'C510223': {'substance_name': 'ALDH1A1 protein, human', 'registry_number': 'EC 1.2.1.36'}, 'D050697': {'substance_name': 'Retinal Dehydrogenase', 'registry_number': 'EC 1.2.1.36'}}
https://ncbi.nlm.nih.gov/pubmed/29938344
PURPOSE: MetXia-P450 is a novel recombinant retroviral vector that encodes the human cytochrome P450 type 2B6 gene (CYP2B6), Escherichia coli lacZ, and neomycin resistance marker genes. Cytochrome P450 enzymes are primarily expressed in the liver and convert the prodrug cyclophosphamide to an active phosphoramide mustard and acrolein. Gene-based delivery of CYP2B6 to the tumor site leads to local prodrug activation and higher concentrations of the active metabolites at the target site.
EXPERIMENTAL DESIGN: MetXia-P450 was directly injected into metastatic cutaneous tumor nodules on days 1 and 2 and nodules biopsied on day 7. Oral cyclophosphamide (100 mg/m(2)) was administered between days 8 and 22. Subsequent cycles of oral cyclophosphamide were repeated for 2 of 4 weeks. Gene transfer levels in biopsy samples were measured by histologic and quantitative PCR analyses. Safety assessments were made using PCR for vector dissemination to the blood after injection and using PCR and serologic analyses to detect replicating virus. Secondary end points included clinical response, toxicity, and evaluation of antitumor immune responses by measurement of carcinoembryonic antigen and 5T4 antibodies.
RESULTS: Twelve patients with breast cancer (n = 9) and melanoma (n = 3) received three dose levels of MetXia-P450 ( approximately 8 x 10(5), approximately 8 x 10(6), and approximately 8 x 10(7) lacZ transferring units/mL). The product was safe and well tolerated. The lacZ transgene was detected in biopsy material by immunohistochemistry in 10 of 12 patients and integrated viral sequences by PCR in 3 of 6 patients. One (8%) patient with breast cancer had a partial response and received 7 months of oral cyclophosphamide. Four (33%) patients had stable disease for > or =3 months and the rest had progressive disease. Preliminary immunologic analyses were suggestive of an antitumor response in two patients (partial response in one patient and stable disease in one patient).
CONCLUSION: MetXia was safe and well tolerated. Gene transfer was detected at all dose levels, and the initial suggestion of an antitumor response indicates that MetXia-P450 should undergo further clinical assessment.
Clin Cancer Res
10.1158/1078-0432.CCR-04-0155
{'D018906': {'substance_name': 'Antineoplastic Agents, Alkylating', 'registry_number': '0'}, 'D002272': {'substance_name': 'Carcinoembryonic Antigen', 'registry_number': '0'}, 'D018396': {'substance_name': 'Mucin-1', 'registry_number': '0'}, 'D003520': {'substance_name': 'Cyclophosphamide', 'registry_number': '8N3DW7272P'}, 'D001189': {'substance_name': 'Aryl Hydrocarbon Hydroxylases', 'registry_number': 'EC 1.14.14.1'}, 'C585599': {'substance_name': 'CYP2B6 protein, human', 'registry_number': 'EC 1.14.14.1'}, 'D065702': {'substance_name': 'Cytochrome P-450 CYP2B6', 'registry_number': 'EC 1.14.14.1'}, 'D010089': {'substance_name': 'Oxidoreductases, N-Demethylating', 'registry_number': 'EC 1.5.-'}}
https://ncbi.nlm.nih.gov/pubmed/15746054
Polymorphisms in drug-metabolizing enzymes and drug transporters contribute to wide and inheritable variability in drug pharmacokinetics, response and toxicity. One of the less well-studied human cytochrome P450s is (CYP)2B6, a homologue of the rodent phenobarbital-inducible CYP2B enzymes. Clinically used drug substrates include cytostatics (cyclophosphamide), HIV drugs (efavirenz and nevirapine), antidepressants (bupropion), antimalarials (artemisinin), anesthetics (propofol) and synthetic opioids (methadone). Contrary to the model polymorphisms of CYP2D6 and CYP2C19, which were discovered by adverse drug reactions, pharmacogenetic study of CYP2B6 was initiated by reverse genetics approaches and subsequent functional and clinical studies. With over 100 described SNPs, numerous complex haplotypes and distinct ethnic frequencies, CYP2B6 is one of the most polymorphic CYP genes in humans. In this review, we summarize general biomolecular and pharmacological features and present a detailed up-to-date description of genetic polymorphisms, including a discussion of recent clinical applications of CYP2B6 pharmacogenetics.
Pharmacogenomics
10.2217/14622416.8.7.743
{'D001189': {'substance_name': 'Aryl Hydrocarbon Hydroxylases', 'registry_number': 'EC 1.14.14.1'}, 'C585599': {'substance_name': 'CYP2B6 protein, human', 'registry_number': 'EC 1.14.14.1'}, 'D065702': {'substance_name': 'Cytochrome P-450 CYP2B6', 'registry_number': 'EC 1.14.14.1'}, 'D010089': {'substance_name': 'Oxidoreductases, N-Demethylating', 'registry_number': 'EC 1.5.-'}}
https://ncbi.nlm.nih.gov/pubmed/17638512
The present study identifies the specific human cytochrome P-450 (CYP) enzymes involved in hydroxylation leading to activation of the anticancer drug cyclophosphamide and its isomeric analogue, ifosphamide. Substantial interindividual variation (4-9-fold) was observed in the hydroxylation of these oxazaphosphorines by a panel of 12 human liver microsomes, and a significant correlation was obtained between these 2 activities (r = 0.85, P < 0.001). Enzyme kinetic analyses revealed that human liver microsomal cyclophosphamide 4-hydroxylation and ifosphamide 4-hydroxylation are best described by a 2-component Michaelis-Menten model composed of both low Km and high Km P-450 4-hydroxylases. To ascertain whether one or more human P-450 enzymes are catalytically competent in activating these oxazaphosphorines, microsomal fractions prepared from a panel of human B-lymphoblastoid cell lines stably transformed with individual P-450 complementary DNAs were assayed in vitro for oxazaphosphorine activation. Expressed CYP2A6, -2B6, -2C8, -2C9, and -3A4 were catalytically competent in hydroxylating cyclophosphamide and ifosphamide. Whereas CYP2C8 and CYP2C9 have the characteristics of low Km oxazaphosphorine 4-hydroxylases, CYP2A6, -2B6, and -3A4 are high Km forms. In contrast, CYP1A1, -1A2, -2D6, and -2E1 did not produce detectable activities. Furthermore, growth of cultured CYP2A6- and CYP2B6-expressing B-lymphoblastoid cells, but not of CYP-negative control cells, was inhibited by cyclophosphamide and ifosphamide as a consequence of prodrug activation to cytotoxic metabolites. Experiments with P-450 form-selective chemical inhibitors and inhibitory anti-P-450 antibodies were then performed to determine the contributions of individual P-450s to the activation of these drugs in human liver microsomes. Orphenadrine (a CYP2B6 inhibitor) and anti-CYP2B IgG inhibited microsomal cyclophosphamide hydroxylation to a greater extent than ifosphamide hydroxylation, consistent with the 8-fold higher activity of complementary DNA-expressed CYP2B6 with cyclophosphamide. In contrast, troleandomycin, a selective inhibitor of CYP3A3 and -3A4, and anti-CYP3A IgG substantially inhibited microsomal ifosphamide hydroxylation but had little or no effect on microsomal cyclophosphamide hydroxylation. By contrast, the CYP2D6-selective inhibitor quinidine did not affect either microsomal activity, while anti-CYP2A antibodies had only a modest inhibitory effect. Overall, the present study establishes that liver microsomal CYP2B and CYP3A preferentially catalyze cyclophosphamide and ifosphamide 4-hydroxylation, respectively, suggesting that liver P-450-inducing agents targeted at these enzymes might be used in cancer patients to enhance drug activation and therapeutic efficacy.
Cancer Res
None
{'D000906': {'substance_name': 'Antibodies', 'registry_number': '0'}, 'D065607': {'substance_name': 'Cytochrome P-450 Enzyme Inhibitors', 'registry_number': '0'}, 'D003520': {'substance_name': 'Cyclophosphamide', 'registry_number': '8N3DW7272P'}, 'D003577': {'substance_name': 'Cytochrome P-450 Enzyme System', 'registry_number': '9035-51-2'}, 'D009966': {'substance_name': 'Orphenadrine', 'registry_number': 'AL805O9OG9'}, 'C104464': {'substance_name': 'CYP3A protein, human', 'registry_number': 'EC 1.14.14.1'}, 'D051544': {'substance_name': 'Cytochrome P-450 CYP3A', 'registry_number': 'EC 1.14.14.1'}, 'D007069': {'substance_name': 'Ifosfamide', 'registry_number': 'UM20QQM95Y'}}
https://ncbi.nlm.nih.gov/pubmed/8242617
BACKGROUND: Doxorubicin and cyclophosphamide (AC) therapy is an effective treatment for early-stage breast cancer. Doxorubicin is a substrate for ABCB1 and SLC22A16 transporters. Cyclophosphamide is a prodrug that requires oxidation to 4-hydroxycyclophosphamide, which yields a cytotoxic alkylating agent. The initial oxidation is catalysed by cytochrome P450 enzymes including CYP2B6, CYP2C9, CYP2C19 and CYP3A5. Polymorphic variants of the genes coding for these enzymes and transporters have been identified, which may influence the systemic pharmacology of the two drugs. It is not known whether this genetic variation has an impact on the efficacy or toxicity of AC therapy.
METHODS: Germ line DNA samples from 230 patients with breast cancer on AC therapy were genotyped for the following SNPs: ABCB1 C1236T, G2677T/A and C3435T, SLC22A16 A146G, T312C, T755C and T1226C, CYP2B6*2, *8, *9, *3, *4 and *5, CYP2C9*2 and *3, CYP3A5*3 and CYP2C19*2. Clinical data on survival, toxicity, demographics and pathology were collated.
RESULTS: A lower incidence of dose delay, indicative of less toxicity, was seen in carriers of the SLC22A16 A146G, T312C, T755C variants. In contrast, a higher incidence of dose delay was seen in carriers of the SLC22A16 1226C, CYP2B6*2 and CYP2B6*5 alleles. The ABCB1 2677A, CYP2B6*2, CYP 2B6*8, CYP 2B6*9, CYP 2B6*4 alleles were associated with a worse outcome.
CONCLUSION: Variant alleles in the ABCB1, SLC22A16 and CYP2B6 genes are associated with response to AC therapy in the treatment of breast cancer.
Br J Cancer
10.1038/sj.bjc.6605587
{'C513055': {'substance_name': 'ABCB1 protein, human', 'registry_number': '0'}, 'D018435': {'substance_name': 'ATP Binding Cassette Transporter, Subfamily B', 'registry_number': '0'}, 'D020168': {'substance_name': 'ATP Binding Cassette Transporter, Subfamily B, Member 1', 'registry_number': '0'}, 'D054316': {'substance_name': 'Biomarkers, Pharmacological', 'registry_number': '0'}, 'D014408': {'substance_name': 'Biomarkers, Tumor', 'registry_number': '0'}, 'D027701': {'substance_name': 'Organic Cation Transport Proteins', 'registry_number': '0'}, 'C467737': {'substance_name': 'SLC22A16 protein, human', 'registry_number': '0'}, 'D004317': {'substance_name': 'Doxorubicin', 'registry_number': '80168379AG'}, 'D003520': {'substance_name': 'Cyclophosphamide', 'registry_number': '8N3DW7272P'}, 'D001189': {'substance_name': 'Aryl Hydrocarbon Hydroxylases', 'registry_number': 'EC 1.14.14.1'}, 'C585599': {'substance_name': 'CYP2B6 protein, human', 'registry_number': 'EC 1.14.14.1'}, 'D065702': {'substance_name': 'Cytochrome P-450 CYP2B6', 'registry_number': 'EC 1.14.14.1'}, 'D010089': {'substance_name': 'Oxidoreductases, N-Demethylating', 'registry_number': 'EC 1.5.-'}}
https://ncbi.nlm.nih.gov/pubmed/20179710
There are a number of reports indicating that CYP2B6*6 (c.516G>T and c.785A>G) is responsible for decreased clearance of efavirenz (EFV), although increased disposition of cyclophosphamide (CPA) in individuals with this polymorphism was observed. Thus, we hypothesized that the effects of the two single nucleotide polymorphisms (SNPs) of CYP2B6*6 on the metabolism of drugs might be considerably different between these two agents. To clarify this possibility, we expressed two major variants of this enzyme, CYP2B6.6 (Q172H and K262R) and CYP2B6.4 (K262R), and investigated metabolic activities of these variants toward EFV and CPA. Kinetic analyses clearly indicated that CYP2B6.4 possessed enhanced metabolic activity toward EFV compared with that of the wild-type enzyme (CYP2B6.1), whereas CPA was metabolized less efficiently by CYP2B6.4 than by CYP2B6.1. On the other hand, CYP2B6.6 showed a completely opposite character, suggesting that Q172H gives inverse effects on metabolic activities of CYP2B6 affected by K262R. Although it is recognized that effects of amino acid change in cytochrome P450 on the metabolic activity depend on substrates, this study revealed SNPs giving an opposite effect on the metabolism of two clinically important drugs currently used. Furthermore, this study provides the first evidence that Q172H can reverse the direction of the effect caused by K262R in CYP2B6 on the metabolism of certain drugs.
Drug Metab Dispos
10.1124/dmd.111.039586
{'D000480': {'substance_name': 'Alkynes', 'registry_number': '0'}, 'D048588': {'substance_name': 'Benzoxazines', 'registry_number': '0'}, 'D003521': {'substance_name': 'Cyclopropanes', 'registry_number': '0'}, 'D003520': {'substance_name': 'Cyclophosphamide', 'registry_number': '8N3DW7272P'}, 'D003577': {'substance_name': 'Cytochrome P-450 Enzyme System', 'registry_number': '9035-51-2'}, 'D006899': {'substance_name': 'Mixed Function Oxygenases', 'registry_number': 'EC 1.-'}, 'D001189': {'substance_name': 'Aryl Hydrocarbon Hydroxylases', 'registry_number': 'EC 1.14.14.1'}, 'C585599': {'substance_name': 'CYP2B6 protein, human', 'registry_number': 'EC 1.14.14.1'}, 'D065702': {'substance_name': 'Cytochrome P-450 CYP2B6', 'registry_number': 'EC 1.14.14.1'}, 'D010089': {'substance_name': 'Oxidoreductases, N-Demethylating', 'registry_number': 'EC 1.5.-'}, 'C098320': {'substance_name': 'efavirenz', 'registry_number': 'JE6H2O27P8'}}
https://ncbi.nlm.nih.gov/pubmed/21821736
AIM: Cisplatin and its analogs are potent antitumor agents. However, their use is restricted by significant variability in tumor response and toxicity. There is a great need to identify genetic markers to predict the most important adverse events and patient outcomes.
MATERIALS & METHODS: We have evaluated the association between polymorphisms in 106 genes involved mainly in xenobiotic metabolism, DNA repair, the cell cycle and apoptosis, and outcomes in 104 ovarian cancer patients receiving cisplatin-cyclophosphamide chemotherapy. Arrayed primer extension technology was used to genotype 228 SNPs.
RESULTS: Ten SNPs in nine genes were found to be associated with one or more of the assessed clinical end points. SNPs in TPMT and NQO1 were significantly associated with progression-free survival. Polymorphisms in ERCC5, RAD52, MUTYH and LIG3 correlated with the occurrence of severe neutropenia. SNPs in NAT2 and EPHX1 were associated with anemia and nephrotoxicity, respectively. A SNP in ADH1C was correlated with complete tumor response.
CONCLUSION: The results obtained suggest that SNPs in different genes involved in drug metabolism can be important in identifying patients at risk for nonresponse to or toxicity from cisplatin-based treatment.
Pharmacogenomics
10.2217/pgs.13.237
{'D054316': {'substance_name': 'Biomarkers, Pharmacological', 'registry_number': '0'}, 'D002945': {'substance_name': 'Cisplatin', 'registry_number': 'Q20Q21Q62J'}}
https://ncbi.nlm.nih.gov/pubmed/24533712
BACKGROUND: Doxorubicin and cyclophosphamide (AC) therapy is an effective treatment for early-stage breast cancer. Doxorubicin is a substrate for ABCB1 and SLC22A16 transporters. Cyclophosphamide is a prodrug that requires oxidation to 4-hydroxycyclophosphamide, which yields a cytotoxic alkylating agent. The initial oxidation is catalysed by cytochrome P450 enzymes including CYP2B6, CYP2C9, CYP2C19 and CYP3A5. Polymorphic variants of the genes coding for these enzymes and transporters have been identified, which may influence the systemic pharmacology of the two drugs. It is not known whether this genetic variation has an impact on the efficacy or toxicity of AC therapy.
METHODS: Germ line DNA samples from 230 patients with breast cancer on AC therapy were genotyped for the following SNPs: ABCB1 C1236T, G2677T/A and C3435T, SLC22A16 A146G, T312C, T755C and T1226C, CYP2B6*2, *8, *9, *3, *4 and *5, CYP2C9*2 and *3, CYP3A5*3 and CYP2C19*2. Clinical data on survival, toxicity, demographics and pathology were collated.
RESULTS: A lower incidence of dose delay, indicative of less toxicity, was seen in carriers of the SLC22A16 A146G, T312C, T755C variants. In contrast, a higher incidence of dose delay was seen in carriers of the SLC22A16 1226C, CYP2B6*2 and CYP2B6*5 alleles. The ABCB1 2677A, CYP2B6*2, CYP 2B6*8, CYP 2B6*9, CYP 2B6*4 alleles were associated with a worse outcome.
CONCLUSION: Variant alleles in the ABCB1, SLC22A16 and CYP2B6 genes are associated with response to AC therapy in the treatment of breast cancer.
Br J Cancer
10.1038/sj.bjc.6605587
{'C513055': {'substance_name': 'ABCB1 protein, human', 'registry_number': '0'}, 'D018435': {'substance_name': 'ATP Binding Cassette Transporter, Subfamily B', 'registry_number': '0'}, 'D020168': {'substance_name': 'ATP Binding Cassette Transporter, Subfamily B, Member 1', 'registry_number': '0'}, 'D054316': {'substance_name': 'Biomarkers, Pharmacological', 'registry_number': '0'}, 'D014408': {'substance_name': 'Biomarkers, Tumor', 'registry_number': '0'}, 'D027701': {'substance_name': 'Organic Cation Transport Proteins', 'registry_number': '0'}, 'C467737': {'substance_name': 'SLC22A16 protein, human', 'registry_number': '0'}, 'D004317': {'substance_name': 'Doxorubicin', 'registry_number': '80168379AG'}, 'D003520': {'substance_name': 'Cyclophosphamide', 'registry_number': '8N3DW7272P'}, 'D001189': {'substance_name': 'Aryl Hydrocarbon Hydroxylases', 'registry_number': 'EC 1.14.14.1'}, 'C585599': {'substance_name': 'CYP2B6 protein, human', 'registry_number': 'EC 1.14.14.1'}, 'D065702': {'substance_name': 'Cytochrome P-450 CYP2B6', 'registry_number': 'EC 1.14.14.1'}, 'D010089': {'substance_name': 'Oxidoreductases, N-Demethylating', 'registry_number': 'EC 1.5.-'}}
https://ncbi.nlm.nih.gov/pubmed/20179710
Several studies have investigated the effects of polymorphisms in the GSTP1, GSTT1, and GSTM1 genes on responsiveness to chemotherapy in breast cancer, but the results have been inconsistent. The aim of this study was to determine the association between polymorphisms of GSTP1, GSTT1, and GSTM1 genes and response to chemotherapy in patients with breast cancer. The relevant studies were retrieved from PubMed, Embase, ISI Web of Knowledge, China National Knowledge Infrastructure, and Wanfang databases. The articles evaluating the correlations between response to chemotherapy and GSTP1, GSTT1, and GSTM1 polymorphisms in breast cancer patients were comprehensively reviewed. Odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated to measure the strength of the associations. These associations were assessed with the χ
Cancer Chemother Pharmacol
10.1007/s00280-016-3173-9
{'C413545': {'substance_name': 'glutathione S-transferase T1', 'registry_number': 'EC 2.5.1.-'}, 'C496556': {'substance_name': 'GSTP1 protein, human', 'registry_number': 'EC 2.5.1.18'}, 'D051549': {'substance_name': 'Glutathione S-Transferase pi', 'registry_number': 'EC 2.5.1.18'}, 'D005982': {'substance_name': 'Glutathione Transferase', 'registry_number': 'EC 2.5.1.18'}, 'C117740': {'substance_name': 'glutathione S-transferase M1', 'registry_number': 'EC 2.5.1.18'}}
https://ncbi.nlm.nih.gov/pubmed/27785604
PURPOSE: Cyclophosphamide and doxorubicin (adjuvant chemotherapy) are commonly used to treat breast cancer patients. Variation in the genes involved in pharmacodynamics and pharmacokinetics of these drugs plays an important role in prediction of drug response and survival. The present study was carried out with an aim to evaluate the variation in all the genes involved in pharmacokinetic and pharmacodynamics pathways of cyclophosphamide and doxorubicin, and correlate specific variants with disease outcome in breast cancer patients from the Malwa region of Punjab.
METHODS: A total of 250 confirmed breast cancer patients were involved in the study. Genotyping was performed on an Illumina Infinium HD assay platform using a Global Screening Array (GSA) microchip. GenomeStudio (Illumina, Inc.) was used for data preprocessing and a p value less than or equal to 5 × 10-8 was considered statistically significant. To rule out the influence of confounding risk factors, a step-wise multivariate regression analysis was carried out to evaluate the association of genotype with overall clinical outcome.
RESULTS: Two gene variants, CYP2C19 (G681A) and ALDH1A1*2 (17 bp deletion), were found to be significantly associated with the disease outcome, including overall survival, recurrence and metastasis, in breast cancer patients on adjuvant therapy. Both these genes are involved in the pharmacokinetics of cyclophosphamide. However, none of the variants in the genes involved in pharmacokinetics and pharmacodynamics of doxorubicin were found to have any significant impact on disease outcome in the studied group.
CONCLUSION: CYP2C19 (G681A) variant and ALDH1A1*2 emerged as two important biomarkers associated with bad outcome in breast cancer patients on adjuvant therapy.
Eur J Clin Pharmacol
10.1007/s00228-018-2505-6
{'D000903': {'substance_name': 'Antibiotics, Antineoplastic', 'registry_number': '0'}, 'D018906': {'substance_name': 'Antineoplastic Agents, Alkylating', 'registry_number': '0'}, 'D014408': {'substance_name': 'Biomarkers, Tumor', 'registry_number': '0'}, 'D004317': {'substance_name': 'Doxorubicin', 'registry_number': '80168379AG'}, 'D003520': {'substance_name': 'Cyclophosphamide', 'registry_number': '8N3DW7272P'}, 'C045793': {'substance_name': 'CYP2C19 protein, human', 'registry_number': 'EC 1.14.14.1'}, 'D065731': {'substance_name': 'Cytochrome P-450 CYP2C19', 'registry_number': 'EC 1.14.14.1'}, 'D000080924': {'substance_name': 'Aldehyde Dehydrogenase 1 Family', 'registry_number': 'EC 1.2.1'}, 'D000444': {'substance_name': 'Aldehyde Dehydrogenase', 'registry_number': 'EC 1.2.1.3'}, 'C510223': {'substance_name': 'ALDH1A1 protein, human', 'registry_number': 'EC 1.2.1.36'}, 'D050697': {'substance_name': 'Retinal Dehydrogenase', 'registry_number': 'EC 1.2.1.36'}}
https://ncbi.nlm.nih.gov/pubmed/29938344
PURPOSE: Veliparib, a PARP inhibitor, demonstrated clinical activity in combination with oral cyclophosphamide in patients with BRCA-mutant solid tumors in a phase I trial. To define the relative contribution of PARP inhibition to the observed clinical activity, we conducted a randomized phase II trial to determine the response rate of veliparib in combination with cyclophosphamide compared with cyclophosphamide alone in patients with pretreated BRCA-mutant ovarian cancer or in patients with pretreated primary peritoneal, fallopian tube, or high-grade serous ovarian cancers (HGSOC).
EXPERIMENTAL DESIGN: Adult patients were randomized to receive cyclophosphamide alone (50 mg orally once daily) or with veliparib (60 mg orally once daily) in 21-day cycles. Crossover to the combination was allowed at disease progression.
RESULTS: Seventy-five patients were enrolled and 72 were evaluable for response; 38 received cyclophosphamide alone and 37 the combination as their initial treatment regimen. Treatment was well tolerated. One complete response was observed in each arm, with three partial responses (PR) in the combination arm and six PRs in the cyclophosphamide alone arm. Genetic sequence and expression analyses were performed for 211 genes involved in DNA repair; none of the detected genetic alterations were significantly associated with treatment benefit.
CONCLUSION: This is the first trial that evaluated single-agent, low-dose cyclophosphamide in HGSOC, peritoneal, fallopian tube, and BRCA-mutant ovarian cancers. It was well tolerated and clinical activity was observed; the addition of veliparib at 60 mg daily did not improve either the response rate or the median progression-free survival.
Clin Cancer Res
10.1158/1078-0432.CCR-14-2565
{'D000970': {'substance_name': 'Antineoplastic Agents', 'registry_number': '0'}, 'D001562': {'substance_name': 'Benzimidazoles', 'registry_number': '0'}, 'C521013': {'substance_name': 'veliparib', 'registry_number': '01O4K0631N'}, 'D003520': {'substance_name': 'Cyclophosphamide', 'registry_number': '8N3DW7272P'}}
https://ncbi.nlm.nih.gov/pubmed/25589624
PURPOSE: Cyclophosphamide and doxorubicin (adjuvant chemotherapy) are commonly used to treat breast cancer patients. Variation in the genes involved in pharmacodynamics and pharmacokinetics of these drugs plays an important role in prediction of drug response and survival. The present study was carried out with an aim to evaluate the variation in all the genes involved in pharmacokinetic and pharmacodynamics pathways of cyclophosphamide and doxorubicin, and correlate specific variants with disease outcome in breast cancer patients from the Malwa region of Punjab.
METHODS: A total of 250 confirmed breast cancer patients were involved in the study. Genotyping was performed on an Illumina Infinium HD assay platform using a Global Screening Array (GSA) microchip. GenomeStudio (Illumina, Inc.) was used for data preprocessing and a p value less than or equal to 5 × 10-8 was considered statistically significant. To rule out the influence of confounding risk factors, a step-wise multivariate regression analysis was carried out to evaluate the association of genotype with overall clinical outcome.
RESULTS: Two gene variants, CYP2C19 (G681A) and ALDH1A1*2 (17 bp deletion), were found to be significantly associated with the disease outcome, including overall survival, recurrence and metastasis, in breast cancer patients on adjuvant therapy. Both these genes are involved in the pharmacokinetics of cyclophosphamide. However, none of the variants in the genes involved in pharmacokinetics and pharmacodynamics of doxorubicin were found to have any significant impact on disease outcome in the studied group.
CONCLUSION: CYP2C19 (G681A) variant and ALDH1A1*2 emerged as two important biomarkers associated with bad outcome in breast cancer patients on adjuvant therapy.
Eur J Clin Pharmacol
10.1007/s00228-018-2505-6
{'D000903': {'substance_name': 'Antibiotics, Antineoplastic', 'registry_number': '0'}, 'D018906': {'substance_name': 'Antineoplastic Agents, Alkylating', 'registry_number': '0'}, 'D014408': {'substance_name': 'Biomarkers, Tumor', 'registry_number': '0'}, 'D004317': {'substance_name': 'Doxorubicin', 'registry_number': '80168379AG'}, 'D003520': {'substance_name': 'Cyclophosphamide', 'registry_number': '8N3DW7272P'}, 'C045793': {'substance_name': 'CYP2C19 protein, human', 'registry_number': 'EC 1.14.14.1'}, 'D065731': {'substance_name': 'Cytochrome P-450 CYP2C19', 'registry_number': 'EC 1.14.14.1'}, 'D000080924': {'substance_name': 'Aldehyde Dehydrogenase 1 Family', 'registry_number': 'EC 1.2.1'}, 'D000444': {'substance_name': 'Aldehyde Dehydrogenase', 'registry_number': 'EC 1.2.1.3'}, 'C510223': {'substance_name': 'ALDH1A1 protein, human', 'registry_number': 'EC 1.2.1.36'}, 'D050697': {'substance_name': 'Retinal Dehydrogenase', 'registry_number': 'EC 1.2.1.36'}}
https://ncbi.nlm.nih.gov/pubmed/29938344
BACKGROUND: Because inheritance is recognized as playing a role in age at menarche and natural menopause, the development of chemotherapy-induced amenorrhea (CIA) might depend on inherited genetic factors; however, studies that explore such a correlation are few and have received scant attention. Given the importance of this topic we conducted a comprehensive genotype study in young women (≤45 years) with early-stage breast cancer.
METHODS: Our approach tested the effect of variant polymorphisms in drug metabolism enzymes (DMEs) using a predesigned pharmacogenomics panel (TaqMan® OpenArray®, Life Technologies GmbH, Darmstadt, Germany) in premenopausal women (n = 50). Patients received contemporary chemotherapy; in all cases a cyclophosphamide-based regimen with a dose of at least 500 mg/m(2) for six cycles. CIA was considered to be present in women with no resumption of menstrual bleeding within 12 months after completion of chemotherapy or goserelin.
RESULTS: Twenty-six patients (52 %) showed CIA during follow-up whereas 24 women (48 %) remained premenopausal. Of all the DMEs studied, only the SLCO1B1*5 (rs4149056) genotype was associated with the development of CIA (P = 0.017). Of the 26 patients who were homozygous for the T/T allele SLCO1B1*5, 18 (69.2 %) developed CIA compared with 8 (30.8 %) of the 22 patients who were heterozygous (C/T allele). The association of heterozygous SLCO1B1*5 allele (OR 0.038; 95%CI: 0.05-0.92) with a lower risk of developing CIA remained significant in a binary logistic regression analysis that include age, SLCO1B1*5 allele variants, and goserelin therapy.
CONCLUSIONS: Patient age and SLCO1B1*5 allele variants predict the likelihood of young women with breast cancer developing CIA.
BMC Cancer
10.1186/s12885-016-2373-3
{'D000970': {'substance_name': 'Antineoplastic Agents', 'registry_number': '0'}, 'D027381': {'substance_name': 'Liver-Specific Organic Anion Transporter 1', 'registry_number': '0'}, 'C503999': {'substance_name': 'SLCO1B1 protein, human', 'registry_number': '0'}, 'D003520': {'substance_name': 'Cyclophosphamide', 'registry_number': '8N3DW7272P'}}
https://ncbi.nlm.nih.gov/pubmed/27234217
BACKGROUND: Genetic risk factors for febrile neutropenia (FN), the major adverse event of perioperative chemotherapy for early breast cancer, remain unclear.
METHODS: This study retrospectively explored pharmacogenetic associations of single nucleotide polymorphisms (SNPs) of the uridine glucuronosyltransferase 2B7 (UGT2B7, rs7668258), glutathione-S-transferase pi 1 (GSTP1, rs1695), and microcephalin 1 (MCPH1, rs2916733) genes with chemotherapy-related adverse events in 102 Japanese women who received epirubicin and cyclophosphamide as perioperative chemotherapy for early breast cancer.
RESULTS: The allele frequencies for all of the SNPs were in concordance with the Hap-Map data of Japanese individuals. Among the 24 patients who had FN at least once during all courses of chemotherapy, 23 had the A/A genotype, and 1 had the A/G genotype of the GSTP1 polymorphism (rs1695, P = 0.001); 23 of the 70 patients with the A/A genotype had FN, as compared with only 1 of the 32 patients with the A/G and G/G genotypes. The genotype distributions of the UGT2B7 and MCPH1 polymorphisms did not differ between the patients who had FN or grade 3/4 neutropenia and those who did not.
CONCLUSION: Among Japanese women who received epirubicin and cyclophosphamide as perioperative chemotherapy for early breast cancer, those with the A/A genotype of the GSTP1 polymorphism (rs1695) were more likely to have FN.
Breast Cancer
10.1007/s12282-014-0547-x
{'D014408': {'substance_name': 'Biomarkers, Tumor', 'registry_number': '0'}, 'D011960': {'substance_name': 'Receptors, Estrogen', 'registry_number': '0'}, 'D015251': {'substance_name': 'Epirubicin', 'registry_number': '3Z8479ZZ5X'}, 'D003520': {'substance_name': 'Cyclophosphamide', 'registry_number': '8N3DW7272P'}, 'C496556': {'substance_name': 'GSTP1 protein, human', 'registry_number': 'EC 2.5.1.18'}, 'D051549': {'substance_name': 'Glutathione S-Transferase pi', 'registry_number': 'EC 2.5.1.18'}, 'C508053': {'substance_name': 'ERBB2 protein, human', 'registry_number': 'EC 2.7.10.1'}, 'D018719': {'substance_name': 'Receptor, ErbB-2', 'registry_number': 'EC 2.7.10.1'}, 'D005472': {'substance_name': 'Fluorouracil', 'registry_number': 'U3P01618RT'}}
https://ncbi.nlm.nih.gov/pubmed/25008867
The glutathione S-transferase (GST) family consists of phase II detoxification enzymes that catalyze the conjugation of toxic substances, such as chemotherapeutic agents, to glutathione. We examined whether GSTT1/GSTT1"null", GSTM1/GSTM1"null" and GSTP1Ile105Ile/GSTP1Ile105Val polymorphisms are associated with different response rates to neoadjuvant chemotherapy in the treatment of stage II and III breast cancer. Forty Brazilian women with invasive ductal adenocarcinoma of the breast submitted to neoadjuvant chemotherapy, using 5-fluorouracil, epirubicin and cyclophosphamide, were genotyped for the GSTT1, GSTM1 and GSTP1 genes. Clinical response was assessed by RECIST criteria. Comparisons were made for the three genes alone and in pairs, as polymorphic and as wild-type combinations and polymorphic/wild-type combinations. We analyzed all possible combinations and their response rate. Patients with the GSTT1/GSTP1105Ile combination were found to have a significantly better response than GSTT1"null"/GSTP1105Val (P = 0.0209) and GSTT1/GSTM1 (P = 0.0376) combinations. Analysis of all possible combinations showed the GSTM1"null" polymorphic genotype to be present in four, and the wild-type GSTP1105Ile in six of the combinations associated with the largest number of responding patients. We found that patients with the GSTT1/GSTP1105Ile wild-type combination had a significantly higher response rate to chemotherapy than patients with the respective polymorphic GSTT1"null"/GSTP1105Val combination or patients with the wild-type GSTT1/GSTM1. The six gene combinations associated with the largest number of responding patients were found to contain the wild-type GSTP1105Ile and the polymorphic-type GSTM1"null". These specific combinations were virtually absent in the combinations with few responding patients.
Genet Mol Res
10.4238/vol9-2gmr726
{'D000970': {'substance_name': 'Antineoplastic Agents', 'registry_number': '0'}, 'D015251': {'substance_name': 'Epirubicin', 'registry_number': '3Z8479ZZ5X'}, 'D003520': {'substance_name': 'Cyclophosphamide', 'registry_number': '8N3DW7272P'}, 'C413545': {'substance_name': 'glutathione S-transferase T1', 'registry_number': 'EC 2.5.1.-'}, 'C496556': {'substance_name': 'GSTP1 protein, human', 'registry_number': 'EC 2.5.1.18'}, 'D051549': {'substance_name': 'Glutathione S-Transferase pi', 'registry_number': 'EC 2.5.1.18'}, 'D005982': {'substance_name': 'Glutathione Transferase', 'registry_number': 'EC 2.5.1.18'}, 'C117740': {'substance_name': 'glutathione S-transferase M1', 'registry_number': 'EC 2.5.1.18'}, 'D005978': {'substance_name': 'Glutathione', 'registry_number': 'GAN16C9B8O'}, 'D005472': {'substance_name': 'Fluorouracil', 'registry_number': 'U3P01618RT'}}
https://ncbi.nlm.nih.gov/pubmed/20568049
Cyclophosphamide (CPA)-based combination treatment has known to be effective for breast cancer, but often causes adverse drug reactions (ADRs). Hence, the identification of patients at risk for toxicity by CPA is clinically significant. In this study, a stepwise case-control association study was conducted using 403 patients with breast cancer who received the CPA combination therapy. A total of 143 genetic polymorphisms in 13 candidate genes (CYP2B6, CYP2C9, CYP2C19, CYP3A4, CYP3A5, ALDH1A1, ALDH3A1, GSTA1, GSTM1, GSTP1, GSTT1, ABCC2 and ABCC4), possibly involved in the activation, metabolism and transport of CPA, were genotyped using 184 cases who developed either > or =grade 3 leukopenia/neutropenia or > or =grade 2 gastrointestinal toxicity and 219 controls who did not show any ADRs throughout the treatment. The association study revealed that one SNP, rs9561778 in ABCC4, showed a significant association with CPA-induced ADRs (Cochran-Armitage trend's P-value=0.00031; odds ratio (OR)=2.06). Subgroup analysis also indicated that the SNP rs9561778 was significantly associated with two major ADR subgroups; gastrointestinal toxicity and leukopenia/neutropenia (Cochran-Armitage trend's P-value=0.00019 and 0.014; OR=2.31 and 1.83). Furthermore, the SNP rs9561778 showed an association with breast cancer patients who were treated with CA(F) drug regimen-induced ADR (Cochran-Armitage trend's P-value=0.00028; OR=3.13). The SNPs in ABCC4 might be applicable in predicting the risk of ADRs in patients receiving CPA combination chemotherapy.
J Hum Genet
10.1038/jhg.2009.79
{'C073492': {'substance_name': 'ABCC4 protein, human', 'registry_number': '0'}, 'D027425': {'substance_name': 'Multidrug Resistance-Associated Proteins', 'registry_number': '0'}, 'D003520': {'substance_name': 'Cyclophosphamide', 'registry_number': '8N3DW7272P'}}
https://ncbi.nlm.nih.gov/pubmed/19696793
PURPOSE: Cyclophosphamide and doxorubicin (adjuvant chemotherapy) are commonly used to treat breast cancer patients. Variation in the genes involved in pharmacodynamics and pharmacokinetics of these drugs plays an important role in prediction of drug response and survival. The present study was carried out with an aim to evaluate the variation in all the genes involved in pharmacokinetic and pharmacodynamics pathways of cyclophosphamide and doxorubicin, and correlate specific variants with disease outcome in breast cancer patients from the Malwa region of Punjab.
METHODS: A total of 250 confirmed breast cancer patients were involved in the study. Genotyping was performed on an Illumina Infinium HD assay platform using a Global Screening Array (GSA) microchip. GenomeStudio (Illumina, Inc.) was used for data preprocessing and a p value less than or equal to 5 × 10-8 was considered statistically significant. To rule out the influence of confounding risk factors, a step-wise multivariate regression analysis was carried out to evaluate the association of genotype with overall clinical outcome.
RESULTS: Two gene variants, CYP2C19 (G681A) and ALDH1A1*2 (17 bp deletion), were found to be significantly associated with the disease outcome, including overall survival, recurrence and metastasis, in breast cancer patients on adjuvant therapy. Both these genes are involved in the pharmacokinetics of cyclophosphamide. However, none of the variants in the genes involved in pharmacokinetics and pharmacodynamics of doxorubicin were found to have any significant impact on disease outcome in the studied group.
CONCLUSION: CYP2C19 (G681A) variant and ALDH1A1*2 emerged as two important biomarkers associated with bad outcome in breast cancer patients on adjuvant therapy.
Eur J Clin Pharmacol
10.1007/s00228-018-2505-6
{'D000903': {'substance_name': 'Antibiotics, Antineoplastic', 'registry_number': '0'}, 'D018906': {'substance_name': 'Antineoplastic Agents, Alkylating', 'registry_number': '0'}, 'D014408': {'substance_name': 'Biomarkers, Tumor', 'registry_number': '0'}, 'D004317': {'substance_name': 'Doxorubicin', 'registry_number': '80168379AG'}, 'D003520': {'substance_name': 'Cyclophosphamide', 'registry_number': '8N3DW7272P'}, 'C045793': {'substance_name': 'CYP2C19 protein, human', 'registry_number': 'EC 1.14.14.1'}, 'D065731': {'substance_name': 'Cytochrome P-450 CYP2C19', 'registry_number': 'EC 1.14.14.1'}, 'D000080924': {'substance_name': 'Aldehyde Dehydrogenase 1 Family', 'registry_number': 'EC 1.2.1'}, 'D000444': {'substance_name': 'Aldehyde Dehydrogenase', 'registry_number': 'EC 1.2.1.3'}, 'C510223': {'substance_name': 'ALDH1A1 protein, human', 'registry_number': 'EC 1.2.1.36'}, 'D050697': {'substance_name': 'Retinal Dehydrogenase', 'registry_number': 'EC 1.2.1.36'}}
https://ncbi.nlm.nih.gov/pubmed/29938344
As judged by findings in preclinical models, determinants of cellular sensitivity to cyclophosphamide and other oxazaphosphorines include two cytosolic aldehyde dehydrogenases, viz., ALDH1A1 and ALDH3A1. Each catalyzes the detoxification of the oxazaphosphorines; thus, cellular sensitivity to these agents decreases as cellular levels of ALDH1A1 and/or ALDH3A1 increase. Of particular clinical relevance may be that stable sublines, relatively insensitive to the oxazaphosphorines due to elevated ALDH1A1 or ALDH3A1 levels, emerged when cultured human tumor cells were exposed only once to a high concentration of one of these agents for 30 to 60 minutes. Whether differences in cellular levels of either enzyme accounts for the clinically-encountered uneven therapeutic effectiveness of the oxazaphosphorines remains to be determined. However, it has already been established that measurable levels of these enzymes are found in some, but not all, tumor types, and that in those tumor types where measurable levels are present, e.g., infiltrating ductal carcinomas of the breast, they vary widely from patient to patient. Potentially useful clinical strategies that might be pursued if it turns out that ALDH1A1 and/or ALDH3A1 are, indeed, clinically operative determinants of cellular sensitivity to the oxazaphosphorines include 1) individualizing cancer chemotherapeutic regimens based, at least in part, on the levels of these enzymes in the malignancy of interest, and 2) sensitizing tumor cells that express relatively large amounts of ALDH1A1 and/or ALDH3A1 to the oxazaphosphorines by preventing the synthesis of these enzymes, e.g., with antisense RNA, or by introducing an agent that directly inhibits the catalytic action of the operative enzyme. Further, the fact that ALDH1A1 and ALDH3A1 are determinants of cellular sensitivity to the oxazaphosphorines provides the rationale for the investigation of two additional strategies with clinical potential, viz., decreasing the sensitivity of vulnerable and essential normal cells, e.g., pluripotent hematopoietic cells, to the oxazaphosphorines by selectively transferring into them the genetic information that encodes 1) ALDH1A1 or ALDH3A1, or 2) a signaling factor, the presence of which would directly or indirectly, stably upregulate the expression of these enzymes.
Curr Pharm Des
None
{'D018906': {'substance_name': 'Antineoplastic Agents, Alkylating', 'registry_number': '0'}, 'D007527': {'substance_name': 'Isoenzymes', 'registry_number': '0'}, 'D010752': {'substance_name': 'Phosphoramide Mustards', 'registry_number': '0'}, 'D003520': {'substance_name': 'Cyclophosphamide', 'registry_number': '8N3DW7272P'}, 'D000444': {'substance_name': 'Aldehyde Dehydrogenase', 'registry_number': 'EC 1.2.1.3'}}
https://ncbi.nlm.nih.gov/pubmed/10469894
PURPOSE: A recent study presented first evidence that a single nucleotide polymorphism (SNP) at codon 388 of fibroblast growth factor receptor 4 (FGFR4) gene, causing a transmembrane domain missense mutation (Gly388Arg), is associated with disease outcome in node-positive breast cancer. This article addresses the clinical relevance of this SNP, FGFR4 genotype, phenotype, and HER2 regarding patient outcome and influence of adjuvant systemic therapy in a substantial primary breast cancer collective (n = 372; median follow-up, 94.5 months).
METHODS: Polymerase chain reaction restriction fragment length polymorphism analysis of germ-line polymorphism was performed in uninvolved lymph nodes; FGFR4 and HER2 expression were assessed immunohistochemically in tissue microarrays.
RESULTS: In 51% of patients, homo- or heterozygous Arg388 allele was present. No correlation existed between FGFR4 genotype and expression or HER2 status. In node-negative patients, FGFR4 genotype was not correlated with disease outcome. In node-positive patients, however, FGFR4 Arg388 was significantly associated with poor disease-free survival (DFS; P = .02) and overall survival (OS; P = .04). Notably, this association seems to be attributable to relatively poor therapy response in Arg388 carriers, reflected in their significantly shorter DFS (P = .02) and OS (P = .045) among patients receiving adjuvant systemic therapy. It is also seen as a significant interaction term in a multivariate proportional hazards model with Arg388 carriers having only about half as much benefit from adjuvant systemic therapy as wild-type carriers.
CONCLUSION: According to this study, FGFR4 Arg388 genotype is a marker for breast cancer progression in patients with adjuvant systemic therapy, particularly chemotherapy, and thus may indicate therapy resistance.
J Clin Oncol
10.1200/JCO.2005.04.8587
{'D014408': {'substance_name': 'Biomarkers, Tumor', 'registry_number': '0'}, 'D005819': {'substance_name': 'Genetic Markers', 'registry_number': '0'}, 'D001120': {'substance_name': 'Arginine', 'registry_number': '94ZLA3W45F'}, 'D051499': {'substance_name': 'Receptor, Fibroblast Growth Factor, Type 4', 'registry_number': 'EC 2.7.10.1'}, 'D005998': {'substance_name': 'Glycine', 'registry_number': 'TE7660XO1C'}}
https://ncbi.nlm.nih.gov/pubmed/16822847
BACKGROUND: In breast cancers, only a minority of patients fully benefit from the different chemotherapy regimens currently in use. Identification of markers that could predict the response to a particular regimen would thus be critically important for patient care. In cell lines or animal models, tumor protein p53 (TP53) plays a critical role in modulating the response to genotoxic drugs. TP53 is activated in response to DNA damage and triggers either apoptosis or cell-cycle arrest, which have opposite effects on cell fate. Yet, studies linking TP53 status and chemotherapy response have so far failed to unambiguously establish this paradigm in patients. Breast cancers with a TP53 mutation were repeatedly shown to have a poor outcome, but whether this reflects poor response to treatment or greater intrinsic aggressiveness of the tumor is unknown.
METHODS AND FINDINGS: In this study we analyzed 80 noninflammatory breast cancers treated by frontline (neoadjuvant) chemotherapy. Tumor diagnoses were performed on pretreatment biopsies, and the patients then received six cycles of a dose-dense regimen of 75 mg/m(2) epirubicin and 1,200 mg/m(2) cyclophosphamide, given every 14 days. After completion of chemotherapy, all patients underwent mastectomies, thus allowing for a reliable assessment of chemotherapy response. The pretreatment biopsy samples were used to determine the TP53 status through a highly efficient yeast functional assay and to perform RNA profiling. All 15 complete responses occurred among the 28 TP53-mutant tumors. Furthermore, among the TP53-mutant tumors, nine out of ten of the highly aggressive basal subtypes (defined by basal cytokeratin [KRT] immunohistochemical staining) experienced complete pathological responses, and only TP53 status and basal subtype were independent predictors of a complete response. Expression analysis identified many mutant TP53-associated genes, including CDC20, TTK, CDKN2A, and the stem cell gene PROM1, but failed to identify a transcriptional profile associated with complete responses among TP53 mutant tumors. In patients with unresponsive tumors, mutant TP53 status predicted significantly shorter overall survival. The 15 patients with responsive TP53-mutant tumors, however, had a favorable outcome, suggesting that this chemotherapy regimen can overcome the poor prognosis generally associated with mutant TP53 status.
CONCLUSIONS: This study demonstrates that, in noninflammatory breast cancers, TP53 status is a key predictive factor for response to this dose-dense epirubicin-cyclophosphamide regimen and further suggests that the basal subtype is exquisitely sensitive to this association. Given the well-established predictive value of complete responses for long-term survival and the poor prognosis of basal and TP53-mutant tumors treated with other regimens, this chemotherapy could be particularly suited for breast cancer patients with a mutant TP53, particularly those with basal features.
PLoS Med
10.1371/journal.pmed.0040090
{'C495901': {'substance_name': 'TP53 protein, human', 'registry_number': '0'}, 'D016159': {'substance_name': 'Tumor Suppressor Protein p53', 'registry_number': '0'}, 'D015251': {'substance_name': 'Epirubicin', 'registry_number': '3Z8479ZZ5X'}, 'D003520': {'substance_name': 'Cyclophosphamide', 'registry_number': '8N3DW7272P'}}
https://ncbi.nlm.nih.gov/pubmed/17388661
!git clone https://github.com/billgreenwald/Pubmed-Batch-Download.git
Cloning into 'Pubmed-Batch-Download'...
remote: Enumerating objects: 199, done.�[K
remote: Counting objects: 100% (9/9), done.�[K
remote: Compressing objects: 100% (9/9), done.�[K
remote: Total 199 (delta 3), reused 0 (delta 0), pack-reused 190�[K
Receiving objects: 100% (199/199), 31.23 MiB | 15.93 MiB/s, done.
Resolving deltas: 100% (100/100), done.
%cd "Pubmed-Batch-Download.git/"
[Errno 20] Not a directory: 'Pubmed-Batch-Download.git/'
/content/drive/MyDrive/Yemaachi_works
!python Pubmed-Batch-Download.git/fetch_pdfs.py -pmids 29938344
python3: can't open file 'Pubmed-Batch-Download.git/fetch_pdfs.py': [Errno 20] Not a directory
parent_path = "/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/"
## list of files in the folder:
variant_annotations = glob(f"{parent_path}/*.tsv")
variant_annotations
['/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_ERCC1.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_MGAT4A.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_DLG5.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_SELE.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_ENOSF1.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_CES1.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_CYP1A1.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_CES1P1.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_MTHFR.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_UMPS.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_EXO1.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_PTEN.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_CYP19A1.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_AREG.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_PTGS2.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_TYMP.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_VEGFA.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_MIR2054.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_HLA-G.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_CDA.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_C18orf56.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_MIR27A.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_SLC22A7.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_REV3L.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_ABCB1.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_variants/all-data_ADCY2.tsv']
### check for the number of variants...
print(f"total number of variants annotation: {len(variant_annotations)}")
total number of variants annotation: 26
## check a sample data....
d = pd.read_csv(variant_annotations[0],sep="\t")
d.head()
|
PharmGKB ID |
Variant |
Literature |
Association |
Significance |
P-Value |
# of Cases |
# of Controls |
Biogeographical Groups |
Paper Discusses |
Pediatric |
More Details |
| 0 |
1448568304 |
rs11615 |
PMID:27995989 |
Genotype GG is associated with decreased respo... |
yes |
= 0.047 |
185 |
NaN |
Unknown |
Efficacy |
False |
Patients with the GG genotype had decreased ov... |
| 1 |
1444934692 |
rs11615 |
PMID:25026457 |
Genotypes AG + GG is associated with decreased... |
yes |
= 0.0238 |
67 |
NaN |
Unknown |
Efficacy |
False |
p-value and OR below for multivariate analysis... |
| 2 |
1448568298 |
rs3212986 |
PMID:27995989 |
Genotype CC are not associated with response t... |
no |
> 0.05 |
185 |
NaN |
Unknown |
Efficacy |
False |
No significant association with response, prog... |
| 3 |
1448568469 |
rs3212986 |
PMID:27995989 |
Genotype CC is not associated with risk of Dru... |
no |
> 0.05 |
185 |
NaN |
Unknown |
Toxicity |
False |
No significant association with global toxicit... |
| 4 |
1448568476 |
rs11615 |
PMID:27995989 |
Genotype GG is not associated with risk of Dru... |
no |
> 0.05 |
185 |
NaN |
Unknown |
Toxicity |
False |
No significant association with global toxicit... |
### concatenate all files loaded...
data_loads = pd.concat((pd.read_csv(file,sep="\t") for file in variant_annotations),ignore_index=True)
### set the data loads
data_loads.head()
|
PharmGKB ID |
Variant |
Literature |
Association |
Significance |
P-Value |
# of Cases |
# of Controls |
Biogeographical Groups |
Paper Discusses |
Pediatric |
More Details |
| 0 |
1448568304 |
rs11615 |
PMID:27995989 |
Genotype GG is associated with decreased respo... |
yes |
= 0.047 |
185 |
NaN |
Unknown |
Efficacy |
False |
Patients with the GG genotype had decreased ov... |
| 1 |
1444934692 |
rs11615 |
PMID:25026457 |
Genotypes AG + GG is associated with decreased... |
yes |
= 0.0238 |
67 |
NaN |
Unknown |
Efficacy |
False |
p-value and OR below for multivariate analysis... |
| 2 |
1448568298 |
rs3212986 |
PMID:27995989 |
Genotype CC are not associated with response t... |
no |
> 0.05 |
185 |
NaN |
Unknown |
Efficacy |
False |
No significant association with response, prog... |
| 3 |
1448568469 |
rs3212986 |
PMID:27995989 |
Genotype CC is not associated with risk of Dru... |
no |
> 0.05 |
185 |
NaN |
Unknown |
Toxicity |
False |
No significant association with global toxicit... |
| 4 |
1448568476 |
rs11615 |
PMID:27995989 |
Genotype GG is not associated with risk of Dru... |
no |
> 0.05 |
185 |
NaN |
Unknown |
Toxicity |
False |
No significant association with global toxicit... |
data_loads.shape
(123, 12)
data_loads.tail()
|
PharmGKB ID |
Variant |
Literature |
Association |
Significance |
P-Value |
# of Cases |
# of Controls |
Biogeographical Groups |
Paper Discusses |
Pediatric |
More Details |
| 118 |
769262755 |
rs1045642 |
PMID:21142915 |
Genotype GG is associated with increased risk ... |
yes |
< 0.033 |
74 |
NaN |
Unknown |
Toxicity |
False |
No significant association was found with risk... |
| 119 |
827817217 |
rs1045642 |
PMID:22026922 |
Genotypes AG + GG are not associated with decr... |
no |
= 0.1605 |
121 |
NaN |
European |
Efficacy |
False |
As measured by overall survival and progressio... |
| 120 |
1185002489 |
rs17160359 |
PMCID:PMC4221105 |
Allele T is associated with increased response... |
not stated |
NaN |
89 |
NaN |
East Asian |
Efficacy |
False |
pfSNP identified 2800 SNPS associated with key... |
| 121 |
1444704172 |
rs4702484 |
PMID:25815774 |
Genotype CC is not associated with decreased o... |
no |
= 0.229 |
265 |
NaN |
Unknown |
Efficacy |
False |
Analyzing the entire cohort of capecitabine mo... |
| 122 |
1444704155 |
rs4702484 |
PMID:25815774 |
Genotype CC is associated with decreased progr... |
no |
= 0.018 |
126 |
NaN |
Unknown |
Efficacy |
False |
This result did not remain statistically signi... |
##### save the variants annontation:
data_loads.to_csv("/content/drive/MyDrive/Yemaachi_works/Capecitabine/variants_annontation.csv",index=False)
clinical_path = "/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/"
clinical_files = glob(f"{clinical_path}/*.tsv")
clinical_files
['/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_rs11615.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_MGAT4A.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_DLG5.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_ABCG2.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_SELE.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_ENOSF1.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_CES1.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_CYP1A1.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_CES1P1.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_MTHFR.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_UMPS.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_EXO1.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_PTEN.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_CYP19A1.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_AREG.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_PTGS2.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_TYMP.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_VEGFA.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_HLA-G.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_CDA.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_MIR27A.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_SLC22A7.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_REV3L.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_ABCB1.tsv',
'/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_clinical/all-data_ADCY2.tsv']
print(f"check for number clinical variants: {len(clinical_files)}")
check for number clinical variants: 25
clinical_data = pd.concat((pd.read_csv(file,sep="\t") for file in clinical_files),ignore_index=True)
clinical_data.head()
|
PharmGKB ID |
Level |
Variant |
Gene |
Molecules |
Type |
Phenotype |
Pediatric |
| 0 |
1445401125 |
3 |
rs11615 |
ERCC1 |
capecitabine; radiotherapy |
Efficacy |
Rectal Neoplasms |
False |
| 1 |
1447990925 |
3 |
rs885036 |
MGAT4A |
bevacizumab; capecitabine; cetuximab; oxaliplatin |
Efficacy |
Colorectal Neoplasms |
False |
| 2 |
1447990913 |
3 |
rs885036 |
MGAT4A |
bevacizumab; capecitabine; oxaliplatin |
Efficacy |
Colorectal Neoplasms |
False |
| 3 |
1444667322 |
3 |
rs2289310 |
DLG5 |
capecitabine; fluorouracil |
Efficacy |
Neoplasm Metastasis |
False |
| 4 |
1444686803 |
4 |
rs2231142 |
ABCG2 |
capecitabine; fluorouracil; leucovorin; oxalip... |
Efficacy |
Colorectal Neoplasms |
False |
clinical_data.tail()
|
PharmGKB ID |
Level |
Variant |
Gene |
Molecules |
Type |
Phenotype |
Pediatric |
| 41 |
1446906439 |
3 |
rs2032582 |
ABCB1 |
capecitabine |
Toxicity |
Colorectal Neoplasms; hand-foot syndrome |
False |
| 42 |
1447964542 |
3 |
rs1128503 |
ABCB1 |
capecitabine |
Toxicity |
Colorectal Neoplasms |
False |
| 43 |
981204466 |
3 |
rs1045642 |
ABCB1 |
capecitabine |
Toxicity |
Neoplasms |
False |
| 44 |
1444667305 |
3 |
rs17160359 |
ABCB1 |
capecitabine; fluorouracil |
Efficacy |
Neoplasm Metastasis |
False |
| 45 |
1444704267 |
4 |
rs4702484 |
ADCY2 |
capecitabine |
Efficacy |
Colorectal Neoplasms |
False |
### check the unique elements
clinical_data.Level.value_counts()
3 43
4 3
Name: Level, dtype: int64
#### clinical annontations for various variants
clinical_data.to_csv("/content/drive/MyDrive/Yemaachi_works/Capecitabine/clinical_annontation.csv",index=False)
%pwd
'/content/drive/My Drive/Yemaachi_works'
%cd "/content/drive/MyDrive/Yemaachi_works/Cyclophosphamide /"
/content/drive/MyDrive/Yemaachi_works/Cyclophosphamide
# data_loads.to_csv("variants_annotations.csv",index=False)
# clinical_data.to_csv("clinical_annotations.csv",index=False)
- Extract all known genes that metabolize with other drugs.
- From drug-genes interaction database.
#### drug extraction for given gene
known_genes = ["CYP2C19","CYP2B6","CYP3A4","CYP2C8","GSTP1","CBR3","NOS3","NQO2","ALDHIA1","TOP2A"]
urlpath = "http://dgidb.org/api/v2/interactions.json?genes="
unmatched_keys = ['searchTerm', 'geneName', 'geneLongName', 'entrezId', 'geneCategories', 'interactions']
def geneDrugInteractions(known_gene = None,urlpath=urlpath):
"""
Args:
input the known gene and urlpath...
return:
dataframe for geneCategories and interactions....
"""
genelink = urlpath + known_gene
print(f"Gene name: {known_gene} urlpath name: {genelink}")
try:
request = requests.get(genelink)
if request.status_code == 200:
json_output = request.json()["matchedTerms"][0]
### gene categories
data_geneCategories = pd.DataFrame(json_output["geneCategories"])
data_geneCategories["geneName"] = json_output["geneName"]
data_geneCategories["geneLongName"] = json_output["geneLongName"]
### gene interaction with drugs
data_interactions = pd.DataFrame(json_output["interactions"])
data_interactions["geneName"] = json_output["geneName"]
data_interactions["geneLongName"] = json_output["geneLongName"]
else:
print(f"request status code: {request.status_code}")
return data_geneCategories , data_interactions
except NameError:
request = requests.get(genelink)
print(f"Not accessible link {request.status_code}")
all_geneCategories = []
all_interactions = []
for gene in known_genes:
#### no information for this gene....
if gene == "ALDHIA1":
continue
else:
data_geneCategories , data_interactions = geneDrugInteractions(known_gene= gene)
### store all geneCategories and interactions
all_geneCategories.append(data_geneCategories)
all_interactions.append(data_interactions)
Gene name: CYP2C19 urlpath name: http://dgidb.org/api/v2/interactions.json?genes=CYP2C19
Gene name: CYP2B6 urlpath name: http://dgidb.org/api/v2/interactions.json?genes=CYP2B6
Gene name: CYP3A4 urlpath name: http://dgidb.org/api/v2/interactions.json?genes=CYP3A4
Gene name: CYP2C8 urlpath name: http://dgidb.org/api/v2/interactions.json?genes=CYP2C8
Gene name: GSTP1 urlpath name: http://dgidb.org/api/v2/interactions.json?genes=GSTP1
Gene name: CBR3 urlpath name: http://dgidb.org/api/v2/interactions.json?genes=CBR3
Gene name: NOS3 urlpath name: http://dgidb.org/api/v2/interactions.json?genes=NOS3
Gene name: NQO2 urlpath name: http://dgidb.org/api/v2/interactions.json?genes=NQO2
Gene name: TOP2A urlpath name: http://dgidb.org/api/v2/interactions.json?genes=TOP2A
#### concatenate all dataframes
all_CategoriesGenes = pd.concat(all_geneCategories,axis=0)
all_interactions_dataframe = pd.concat(all_interactions,axis=0)
all_CategoriesGenes.head()
|
id |
name |
geneName |
geneLongName |
| 0 |
d3ec2631e0b2434b9dcc008e793d3fa5 |
DRUGGABLE GENOME |
CYP2C19 |
CYTOCHROME P450 FAMILY 2 SUBFAMILY C MEMBER 19 |
| 1 |
430d0ae401ac4c05ae4a1ad4bee6f23d |
DRUG METABOLISM |
CYP2C19 |
CYTOCHROME P450 FAMILY 2 SUBFAMILY C MEMBER 19 |
| 2 |
e5da6843ba1a43ef9988ba09f2701975 |
CYTOCHROME P450 |
CYP2C19 |
CYTOCHROME P450 FAMILY 2 SUBFAMILY C MEMBER 19 |
| 3 |
4866b8ad-3c3e-4c04-802f-587f697212db |
ENZYME |
CYP2C19 |
CYTOCHROME P450 FAMILY 2 SUBFAMILY C MEMBER 19 |
| 0 |
d3ec2631e0b2434b9dcc008e793d3fa5 |
DRUGGABLE GENOME |
CYP2B6 |
CYTOCHROME P450 FAMILY 2 SUBFAMILY B MEMBER 6 |
all_interactions_dataframe.head()
|
interactionId |
interactionTypes |
drugName |
drugConceptId |
sources |
pmids |
score |
geneName |
geneLongName |
| 0 |
86e8c643-8a82-49c6-b989-177943ef923e |
[] |
CHEMBL372797 |
chembl:CHEMBL372797 |
[DTC] |
[] |
0.14 |
CYP2C19 |
CYTOCHROME P450 FAMILY 2 SUBFAMILY C MEMBER 19 |
| 1 |
b7e6618a-2a5a-42b6-a056-4f3689c756da |
[] |
CHEMBL406845 |
chembl:CHEMBL406845 |
[DTC] |
[] |
0.00 |
CYP2C19 |
CYTOCHROME P450 FAMILY 2 SUBFAMILY C MEMBER 19 |
| 2 |
8ecda335-6ba6-4fc4-9c55-7f1099d970e8 |
[] |
DUP-697 |
chembl:CHEMBL42485 |
[DTC] |
[] |
0.05 |
CYP2C19 |
CYTOCHROME P450 FAMILY 2 SUBFAMILY C MEMBER 19 |
| 3 |
92a30f69-bcc6-4ba7-9fbc-534ecaa91c41 |
[] |
BIOCHANIN |
chembl:CHEMBL131921 |
[DTC] |
[] |
0.02 |
CYP2C19 |
CYTOCHROME P450 FAMILY 2 SUBFAMILY C MEMBER 19 |
| 4 |
b0cb9f03-b1cd-487b-aaec-7f5bb37a717a |
[] |
METARAMINOL |
chembl:CHEMBL1201319 |
[DTC] |
[22931300] |
0.04 |
CYP2C19 |
CYTOCHROME P450 FAMILY 2 SUBFAMILY C MEMBER 19 |
print(f"shape of categories dataframe: {all_CategoriesGenes.shape}")
print(f"shape of interactions dataframe: {all_interactions_dataframe.shape}")
shape of categories dataframe: (28, 4)
shape of interactions dataframe: (1623, 9)
##### save all data
all_CategoriesGenes.to_csv("all_CategoriesGenes.csv",index=False)
all_interactions_dataframe.to_csv("all_interactions_dataframe.csv",index= False)
%pwd
'/content/drive/My Drive/Yemaachi_works'
Retrieve Gene info using NCBI API:
- Retrieve the frequency data for a given gene using the define gene id in NCBI
@limits(calls=1, period=1) # Only one call per second
def get_gene_loc(gene_id: str) -> List[Any]:
'''
Return chromosome id, start and stop positions for gene_id
'''
esum_url=(f'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/'
f'esummary.fcgi?db=gene&id={gene_id}&format=json')
print (f'esummary url: {esum_url}')
res = requests.get(esum_url)
if res.status_code != 200:
raise("Failed to get gene information")
data = res.json()
# First, verify that result contains location data
if ('result' not in data or gene_id not in data['result'] or
'genomicinfo' not in data['result'][gene_id]):
raise("Genomic information is not avaible for this gene")
# Extract and return location data
loc = data['result'][gene_id]['genomicinfo'][0]
chraccver = loc['chraccver']
chrstart = int(loc['chrstart'])
chrstop = int(loc['chrstop'])
# If the gene is on the opposite strand of the reference
# sequence (e.g. TP53), chrstart is larger than chrstop.
# We need to swap them to make sure chrstart < chrstop.
if chrstart > chrstop:
chrstart, chrstop = chrstop, chrstart
return (chraccver, chrstart, chrstop)
#### set the gene id:
gene = "TYMS"
gene_id = "7298"
chraccver, chrstart, chrstop = get_gene_loc(gene_id)
print (f'gene id: {gene_id}, chr: {chraccver}, '
f'start: {chrstart}, stop: {chrstop}.')
esummary url: https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esummary.fcgi?db=gene&id=7298&format=json
gene id: 7298, chr: NC_000018.10, start: 657652, stop: 673577.
def get_next_interval_start(result: dict) -> int:
'''
Return the start position of the next search interval
'''
# Collect stop positions of all 250 variations from the response.
stops = []
for k in result.keys():
length, start = k.split('@')
stops.append(int(length) + int(start))
# The next search interval starts just after the last variant's stop position.
return max(stops) + 1
@limits(calls=1, period=1) # Only one call per second
def get_freq_by_interval(seq_id: str, start: int, stop: int) -> None:
'''
Recursively retrieve frequency data from the overlapping_frequency_records
API service for a given sequence interval.
'''
api_url = (f'https://api.ncbi.nlm.nih.gov/variation/v0/interval/'
f'{seq_id}:{start}:{stop - start + 1}'
f'/overlapping_frequency_records')
print (api_url)
res = requests.get(api_url)
# A global variable that allows for accumulating results from
# recursive calls. It must be reset before each external call
# of get_freq_by_interval
global coll
# Check status_code to decide what to do next
if res.status_code == 200:
# We got all we asked for. Save the result and return.
coll.update(res.json()['results'])
return
elif res.status_code == 206:
# There are more data than the service can return.
# We should save the result, and call the service again with
# the next interval.
coll.update(res.json()['results'])
print (f'Accumulated result size: {len(coll)}')
# Delay the call for 1 second to not exceed the rate limit.
time.sleep(1)
get_freq_by_interval(seq_id, get_next_interval_start(coll), stop)
elif res.status_code >= 400:
raise (f'API request returned with error code {res.status_code}\n'
f'Request: {api_url}\n'
f'Response: {res.json()}')
else:
raise(f'Unexpected return code: {res.status_code}')
# Collect results from get_freq_by_interval
coll = {}
get_freq_by_interval(chraccver, chrstart, chrstop)
print (f'Final result: {len(coll)}')
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:657652:15926/overlapping_frequency_records
Accumulated result size: 250
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:658213:15365/overlapping_frequency_records
Accumulated result size: 500
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:658946:14632/overlapping_frequency_records
Accumulated result size: 750
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:659877:13701/overlapping_frequency_records
Accumulated result size: 1000
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:660803:12775/overlapping_frequency_records
Accumulated result size: 1250
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:661773:11805/overlapping_frequency_records
Accumulated result size: 1500
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:662532:11046/overlapping_frequency_records
Accumulated result size: 1750
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:663351:10227/overlapping_frequency_records
Accumulated result size: 2000
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:664266:9312/overlapping_frequency_records
Accumulated result size: 2250
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:665073:8505/overlapping_frequency_records
Accumulated result size: 2500
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:665986:7592/overlapping_frequency_records
Accumulated result size: 2750
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:666718:6860/overlapping_frequency_records
Accumulated result size: 3000
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:667047:6531/overlapping_frequency_records
Accumulated result size: 3250
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:667370:6208/overlapping_frequency_records
Accumulated result size: 3500
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:667601:5977/overlapping_frequency_records
Accumulated result size: 3750
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:668180:5398/overlapping_frequency_records
Accumulated result size: 4000
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:668890:4688/overlapping_frequency_records
Accumulated result size: 4250
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:669630:3948/overlapping_frequency_records
Accumulated result size: 4500
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:670284:3294/overlapping_frequency_records
Accumulated result size: 4750
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:671087:2491/overlapping_frequency_records
Accumulated result size: 5000
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:671911:1667/overlapping_frequency_records
Accumulated result size: 5250
https://api.ncbi.nlm.nih.gov/variation/v0/interval/NC_000018.10:672793:785/overlapping_frequency_records
Final result: 5498
Convert pdf to text:
### install the library for converting the text:
!pip install pdfminer
Collecting pdfminer
Downloading pdfminer-20191125.tar.gz (4.2 MB)
�[K |████████████████████████████████| 4.2 MB 5.5 MB/s
�[?25hCollecting pycryptodome
Downloading pycryptodome-3.11.0-cp35-abi3-manylinux2010_x86_64.whl (1.9 MB)
�[K |████████████████████████████████| 1.9 MB 36.4 MB/s
�[?25hBuilding wheels for collected packages: pdfminer
Building wheel for pdfminer (setup.py) ... �[?25l�[?25hdone
Created wheel for pdfminer: filename=pdfminer-20191125-py3-none-any.whl size=6140093 sha256=c7c66819d38da571230a0cf1fe3aad4983f8b39b3b9773fc9c9bfe6579b952ca
Stored in directory: /root/.cache/pip/wheels/e3/5e/f4/d210b46e9e4a28229ea070ed5b3efa92c3c29d1a7918dd4b97
Successfully built pdfminer
Installing collected packages: pycryptodome, pdfminer
Successfully installed pdfminer-20191125 pycryptodome-3.11.0
%ls -sh "/content/drive/MyDrive/Yemaachi_works/Capecitabine"
total 5.0M
512 all-data_clinical_annotation.gsheet
5.5K all-data_clinical_annotation.tsv
512 all-data_variant_annotation.gsheet
275K all-data_variant_annotation.tsv
3.0K all_new_variants.csv
512 all_new_variants.gsheet
34K automatic_annotations-PA448771.tsv
4.0K �[0m�[01;34mcapecitabine_clinical�[0m/
512 'CAPECITABINE_genes (1).gsheet'
5.0K CAPECITABINE_genes.csv
512 CAPECITABINE_genes.gsheet
123K capecitabine_genes_paper_summary.csv
512 capecitabine_genes_paper_summary.gsheet
512 Capecitabine.gsheet
512 'CAPECITABINE_interactions_claims (1).gsheet'
11K CAPECITABINE_interactions_claims.csv
512 CAPECITABINE_interactions_claims.gsheet
4.0K �[01;34mcapecitabine_variants�[0m/
4.0K clinical_annontation.csv
512 clinical_annontation.gsheet
897K colorectal_cancer.pdf
3.6M gene_info_id
512 'variants_annontation (1).gsheet'
61K variants_annontation.csv
512 variants_annontation.gsheet
#### set the pdf file to text:
pdf_path = "/content/drive/MyDrive/Yemaachi_works/Capecitabine/colorectal_cancer.pdf"
##### convert pdf file into raw text for mining
from io import StringIO
from pdfminer.converter import TextConverter
from pdfminer.layout import LAParams
from pdfminer.pdfdocument import PDFDocument
from pdfminer.pdfinterp import PDFResourceManager, PDFPageInterpreter
from pdfminer.pdfpage import PDFPage
from pdfminer.pdfparser import PDFParser
def convert_pdf_to_text(file_path: str = None):
output_string = StringIO()
with open(file_path, 'rb') as in_file:
parser = PDFParser(in_file)
doc = PDFDocument(parser)
rsrcmgr = PDFResourceManager()
device = TextConverter(rsrcmgr, output_string, laparams=LAParams())
interpreter = PDFPageInterpreter(rsrcmgr, device)
for page in PDFPage.create_pages(doc):
interpreter.process_page(page)
data = output_string.getvalue()
print(data)
return data
### convert the pdf into text for mining.
text = convert_pdf_to_text(file_path=pdf_path)
Cancer Chemotherapy and Pharmacology
https://doi.org/10.1007/s00280-017-3478-3
ORIGINAL ARTICLE
DPYD*2A and MTHFR C677T predict toxicity and efficacy, respectively,
in patients on chemotherapy with 5-fluorouracil for colorectal cancer
Noor Ahmed Nahid1 · Mohd Nazmul Hasan Apu1 · Md. Reazul Islam1 · Samia Shabnaz1 ·
Surid Mohammad Chowdhury1 · Maizbha Uddin Ahmed1 · Zabun Nahar2 · Md. Siddiqul Islam1 ·
Mohammad Safiqul Islam3
· Abul Hasnat1
Received: 8 July 2017 / Accepted: 8 November 2017
© Springer-Verlag GmbH Germany, part of Springer Nature 2017
Abstract
Background Significant inter-individual variation in the sensitivity to 5-fluorouracil (5-FU) represents a major therapeutic
hindrance either by impairing drug response or inducing adverse drug reactions (ADRs). This study aimed at exploring
the cause behind this inter-individual alterations in consequences of 5-fluorouracil-based chemotherapy by investigating
the effects of DPYD*2A and MTHFR C677T polymorphisms on toxicity and response of 5-FU in Bangladeshi colorectal
cancer patients.
Methods Colorectal cancer patients (n = 161) receiving 5-FU-based chemotherapy were prospectively enrolled. DPYD
and MTHFR polymorphisms were assessed in peripheral leukocytes. Multivariate analyses were applied to evaluate which
variables could predict chemotherapy-induced toxicity and efficacy.
Results Multivariate analyses showed that DPYD*2A polymorphism was a predictive factor (P = 0.023) for grade 3 and grade
4 5-fluorouracil-related toxicities. Although MTHFR C677T polymorphism might act as forecasters for grade 3 or grade 4
neutropenia, diarrhea, and mucositis, this polymorphism was found to increase significantly (P = 0.006) the response of 5-FU.
Conclusion DPYD*2A and MTHFR C677T polymorphisms could explain 5-FU toxicity or clinical outcome in Bangladeshi
colorectal patients.
Keywords Colorectal cancer · 5-Fluorouracil · DPYD · MTHFR · Toxicity · Response
Introduction
Colorectal cancer (CRC) is the third most common cancer
in men and the second most common cancer in women and
the fourth leading cause of cancer-related deaths world-
wide [1]. 5-Fluorouracil (5-FU) is the most commonly used
chemotherapeutic agent for the treatment of CRC either as a
monotherapy or in combination with other chemotherapeutic
drugs and it is central to all chemotherapeutic combinations
* Mohammad Safiqul Islam
research_safiq@yahoo.com
1 Department of Clinical Pharmacy and Pharmacology,
Faculty of Pharmacy, University of Dhaka, Dhaka 1000,
Bangladesh
2 Department of Pharmacy, University of Asia Pacific, Dhaka,
Bangladesh
3 Department of Pharmacy, Noakhali Science and Technology
University, Sonapur, Noakhali 3814, Bangladesh
for CRC treatment [2–11]. Although clinical trials provided
evidence of efficacy and safety of 5-fluorouracil at usual
doses in populations, some patients showed a wide variation
in the response and even adverse effects [11–13]. The use of
5-FU is burdened by a number of serious toxicities including
nausea, diarrhea, mucositis, myelosuppression, dermatologi-
cal toxicities, neurological toxicities, etc. and these toxicities
may even become very fatal to cause death to the patients
[11, 14]. A meta-analysis of 1219 CRC patients revealed that
the overall proportion of grade 3–4 hematologic toxicity was
4–31% and non-hematologic toxicity 13–14% for patients
assigned to 5-FU [15].
To act as an anticancer drug at first, 5-FU is converted
into cytotoxic metabolites and only 1–3% of the adminis-
tered 5-FU is converted into it. Approximately, 80% of the
administered 5FU is degraded by the metabolic enzymes
with the remaining portion being excreted directly in the
urine [16–18]. Here, the catabolizing enzyme is the dihy-
dropyrimidine dehydrogenase (DPYD), which is responsible
Vol.:(0123456789)1 3�
Cancer Chemotherapy and Pharmacology
for converting 5FU into dihydrofluorouracil (FDHU). This
FDHU is further converted into metabolites that are then
excreted through urine [19]. Thus, the deficiency to this
enzyme would result in 5-FU accumulation in the blood,
which may induce severe life-threatening toxicities [20,
21]. Various clinical studies revealed that single nucleotide
polymorphism (SNP) may reduce the catabolic activity of
DPYD enzyme, causing 5-FU accumulation in the blood,
increasing the risk for toxicities [18, 22]. The DPYD activ-
ity due to genetic polymorphisms varies widely in different
populations [23].
Methylenetetrahydrofolatereductase (MTHFR) is another
gene of interest in this study as the activity of this enzyme
can modulate tumor response to 5-FU. The most impor-
tant mechanism for anticancer effects of 5-FU involves
the formation of a complex of 5-fluoro-2-deoxyuridine-
5-monophosphate (5FdUMP), thymidylate synthase (TS),
and 5, 10-methylenetetrahydrofolate (5, 10-methylene-THF),
thereby inhibiting TS activity and, ultimately, inhibiting
DNA synthesis [24, 25]. MTHFR catalyzes the conver-
sion of 5,10-methylene-THF to 5-methyltetrahydrofolate
(5-methyl-THF), and thus the MTHFR C677T polymor-
phism, which may decrease the activity of MTHFR leads
to an accumulation of 5,10-methylene-THF. This excess
5,10-methylene-THF increases the stability of the ternary
complex, facilitating the antitumor effects [25–27].
In this study, we have tried to determine whether DPYD
and MTHFR polymorphisms can induce toxicities in Bang-
ladeshi colorectal patients who were treated with 5-FU-
based combined chemotherapy. This study was also designed
to establish the relationship between MTHFR polymorphism
and tumor response to 5-FU.
Materials and methods
Subject selection
A total of 161 patients histologically proven with colo-
rectal carcinoma were recruited prospectively (i.e., with-
out prior knowledge of the study outcomes—toxicity and
tumor response) from the National Institute of Cancer
Research & Hospital (NICRH), Dhaka, Bangladesh. Ethical
approval was obtained from the National Institute of Cancer
Research & Hospital (NICRH) Ethics Committee. Written
consents for participating in this study were obtained from
all patients. All patients received either FOLFOX (5-fluo-
rouracil, oxaliplatin, and folinic acid) or FOLFIRI (5-fluo-
rouracil, irinotecan, and folinic acid) therapy. For inclusion
in the study, patients had to fulfil the following criteria: (1)
histologically proven colorectal adenocarcinoma with bidi-
mensionally measurable disease; (2) World Health Organiza-
tion performance status < 3; (3) any previous chemotherapy
completed ≥ 6 months ago; (4) life expectancy > 3 months;
(5) adequate hematological and cardiac function; (6) liver
function (serum bilirubin ≤ 2.0 mg/dl, aspartate aminotrans-
ferase (AST) ≤ 3X upper limit of normal value or up to 5X
the upper limit of normal value for patients with liver metas-
tasis); and (7) renal function (serum creatinine ≤ 2.0 mg/
dl). Pre-treatment evaluation included a complete physi-
cal examination done within 2 weeks before the entry into
the study. Pre-treatment evaluation includes—(1) baseline
patient demographics (age, sex, and ethnicity) and medical
history; (2) performance status evaluation; (3) diagnosis of
tumor and staging; (4) computed tomography (CT) scan-
ning of the abdomen and pelvis (chest or any other region if
metastasis was suspected or previously detected); (5) current
chemotherapy regimen (drug and dosing regimen); (6) base-
line blood analyses; and (7) CBC count with leukocyte dif-
ferential, platelet count. All patients were monitored for liver
and renal function, complete blood count, and also assessed
for treatment tolerance before each chemotherapy cycle.
The patients with their guardians were interviewed in the
presence of expert physicians to obtain their demographic
particulars and other physical conditions. We recruited 161
patients for toxicity assessment, but for response evaluation,
data were available for 139 patients, and thus, we excluded
22 patients from response evaluation part of this study.
Patients having a previous history of any invasive malig-
nancy and those who refused to give consent and share their
data were excluded from this study. The study was com-
pleted in accordance with the Declaration of Helsinki and
its further amendments (adopted by the 18th WMA general
assembly, Helsinki, Finland, June 1964 and last amendment
in Seoul, South Korea on October 2008) [28].
Toxicity assessment
For each patient, the maximum observed toxicity grade was
recorded for each toxicity pattern—neutropenia, thrombo-
cytopenia, anemia, leukopenia, nausea, vomiting, mucosi-
tis, diarrhea, dermatological toxicity, and neurological
toxicity. All the types of toxicities were graded according
to the Common Terminology Criteria for Adverse Events
(CTCAE v3.0) [29]. The role of DPYD and MTHFR gene
polymorphisms on 5-FU-induced toxicities was assessed in
this study.
Response evaluation
Tumor response to 5-FU was estimated according to the
Response Evaluation Criteria in Solid Tumors (RECIST)
[30]. We included the patients who had measurable disease
according to RECIST. We recorded complete response
(CR) if there was a disappearance of tumor for at least
4 weeks; partial response (PR) if there was at least a 30%
1 3�Cancer Chemotherapy and Pharmacology
decrease on the longest diameter of tumor for more than
4 weeks; progressive disease (PD) if there was at least a
20% increase on the longest diameter of tumor; and stable
disease (SD) if there was neither sufficient shrinkage to
qualify for partial response nor sufficient increase to qual-
ify for progressive disease [30]. We included 139 patients
of total 161 patients for whom clinical data were available,
to evaluate whether MTHFR polymorphism can modulate
tumor response to 5-FU.
Genotype analysis
Genetic studies were done in the Pharmacogenetics and
Pharmacokinetics Laboratory of Department of Clini-
cal Pharmacy and Pharmacology, University of Dhaka,
Bangladesh. Genomic DNA was extracted from blood
samples of 161 colorectal cancer patients by using meth-
ods as described by Islam et al. [31]. To facilitate the
accurate genotyping of the patient’s DNA samples for
the DPYD*2A or DPYD c.1905+1G>A (rs3918290) and
MTHFR C677T (rs1801133) single nucleotide polymor-
phisms (SNPs), Polymerase Chain Reaction–Restriction
Fragment Length Polymorphism (PCR–RFLP) method
was employed. We used HpyCH4IV and Hinf1 to digest
DPYD*2A and MTHFR C677T PCR products, respec-
tively. The subsequent digestion or lack of digestion, of
PCR amplification product due to the presence or absence
of an SNP within the restriction enzyme recognition site
allowed for accurate and reliable genotyping and the con-
sequent determination of SNP frequencies within a sample
cohort (Figs. 1, 2). The classification of an SNP genotype
as ‘wild type’ or ‘variant’ was done according to accepted
nomenclature and the relevant reference sequences avail-
able from the National Centre for Biotechnological Infor-
mation (NCBI) Entrez Nucleotides Database (http://www.
ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide). All
mutant homozygous and 30% of heterozygotes were ana-
lyzed twice to confirm the genotyping.
Statistical analysis
Associations of the genotypes with the response and tox-
icities of chemotherapy were estimated by computing odds
ratios (ORs) and 95% confidence intervals (CIs) from mul-
tivariate logistic regression with adjustment for age, sex,
BMI, area of residence, ECOG stages, and type of drug
regimen. Differences in the demographic and clinicopatho-
logical characteristics of patients with and without toxicity
and response were tested using Fisher’s exact test (discrete
variables). All statistical analyses were done applying the
SPSS software, version 17.0.
Fig. 1 Restriction endonuclease (HpyCHIV4) digestion fragment of
DPYD:IVS14+1G>A (rs3918290). NH normal homozygote/GG gen-
otype, HE heterozygote/ GA genotype
Fig. 2 Restriction endonuclease (HinfI) digestion fragment of
MTHFR C677T (rs1801133). NH normal homozygote/CC genotype,
HE heterozygote/CT genotype, MH mutant homozygote/TT geno-
type)
1 3�
Results
Clinical data
A total of 161 clinically diagnosed with colorectal carci-
noma, patients were included in this study. The sociode-
mographic and clinicopathological characteristics of the
patients are given in Table 1. The median age of the total 161
patients (97 male, 64 female and 87 colon cancer, and 74 rec-
tum cancer) was 47, ranging from 22 to 75 years and about
10% patients had underweight having BMI less than 18.5 kg/
m2. The patients were treated either by FOLFOX (42.9%)
or by FOLFIRI (57.1%) therapies. During chemotherapy,
Table 1 Characteristics of the patients
Characteristics of the patients
Age (years)
Range
Median
< 45
45–60
> 60
Sex
Male
Female
Body Mass Index (BMI)
< 18.5
18.5–30
Primary tumor site
Colon
Rectum
Dwelling
Rural
Urban
WHO performance status
0
1
2
3
Chemotherapy regimen
5-Fluorouracil, Oxaliplatin, Folinic acid (FOLFOX)
5-Fluorouracil, Irinotecan, Folinic acid (FOLFIRI)
Clinical T-stage
T2
T3
T4
Clinical N-stage
N0
N1
N2
n = 161 (%)
25–75
47
73 (45.34)
53 (32.9)
35 (21.8)
97 (60.2)
64 (39.8)
16 (9.94)
145 (90.06)
87 (54)
74 (46)
88 (54.7)
73 (45.3)
56 (34.78)
73 (45.34)
25 (15.5)
7 (5)
69 (42.9)
92 (57.1)
98 (60.87)
39 (24.22)
24 (14.91)
75 (46.58)
49 (30.43)
37 (22.98)
Cancer Chemotherapy and Pharmacology
78 patients (48.4%) develop grade 3 or grade 4 toxicities of
various types including anemia, neutropenia, thrombocyto-
penia, leukopenia, mucositis, dermatological toxicities, diar-
rhea, nausea, vomiting, and neurological toxicities. Clinical
data for evaluating response were available for 139 patients.
Thus, these 139 patients were included for response evalu-
ation as a part of this study. Out of 139 patients, 81 patients
(58.3%) responded and 58 patients (41.7%) did not respond
to the treatment. The patients received 10–12 cycles of
chemotherapy in average.
Description of analyzed genotypes
Most of the patients (n = 153, about 95%) exhibited wild-
type (wt, G/G) genotype and only eight patients (5%)
were heterozygous (G/A) and no patient was detected as a
homozygous mutant for DPYD*2A polymorphism. Thus,
in case of Bangladeshi colorectal cancer patients, we found
frequency of 2.5% for minor allele of DPYD*2A poly-
morphism. In case of MTHFR C677T polymorphism, 112
patients (69.6%) had C/C, 36 patients (22.3%) exhibited
C/T, and 13 patients (8.1%) showed T/T genotype. Here,
the allelic frequency was 20.5% for the minor allele.
Impact of DPYD and MTHFR gene polymorphisms
on toxicity
Various factors such as age, sex, BMI, performance status,
stage, and grade of the tumor were not significantly associ-
ated with toxicity except N-stage (Table 2). Of the total 161
cases, 21.7% suffered from anemia, 28% from neutropenia,
8% from thrombocytopenia, 10% from leukopenia, 29.2%
from diarrhea, 13% from nausea, 15.5% from vomiting, 13%
from dermatological toxicities, 5.6% from mucositis, and
1.2% from neurological toxicities.
DPYD*2A polymorphism was significantly associ-
ated (P = 0.023) with grade 3 and grade 4 toxicities. About
46.4% patients having G/G genotype suffered from grade 3
or grade 4 toxicities, whereas this percentage was increased
to 87.5 when considering the G/A genotype carrier patients.
This polymorphism is significantly associated with grade 3
and grade 4 anemia (adjusted OR 4.7; 95% CI 1.06–20.96;
P = 0.042), neutropenia (OR 6.47; 95% CI 1.37–30.51,
P = 0.018), thrombocytopenia (adjusted OR 8.08; 95% CI
1.00–65.15; P = 0.050), nausea (adjusted OR 10.06; 95%
CI 1.65–61.26; P = 0.012), and diarrhea (adjusted OR 5.76,
95% CI 1.24–26.77, P = 0.026) when G/A genotype was
compared to G/G genotype (Table 3). In most of the cases,
toxicity occurred after the first or second cycle of chemo-
therapy in carriers of mutant genotypes. All the patients with
DPYD*2A, polymorphism had at least one dose modifica-
tion and only four patients were able to complete the 12
cycles.
1 3�Cancer Chemotherapy and Pharmacology
Table 2 Correlations between clinicopathological features and toxicity status in 161 colorectal cancer patients
Characteristics of the patients
Toxicities Grade 3 + 4,
n = 78 (%)
Total cases, n = 161
(%)
Toxicities Grade ≤ 2,
n = 83 (%)
P value
32 (41)
30 (38.5)
16 (20.5)
43 (55.1)
35 (44.9)
9 (11.5)
69 (88.5)
46 (59)
32 (41)
47 (60.3)
31 (39.7)
12 (15.4)
48 (61.5)
12 (15.4)
6 (7.7)
35 (44.9)
43 (55.1)
44 (56.4)
23 (29.5)
11 (14.1)
45 (57.7)
19 (24.4)
14 (17.9)
87 (54)
74 (46)
97 (60.2)
64 (39.8)
88 (54.7)
73 (45.3)
16 (9.3)
145 (90.7)
73 (45.3)
53 (32.9)
35 (21.8)
Age (years)
< 45
45–60
> 60
Sex
Male
Female
Body Mass Index (BMI)
<18.5
18.5–30
Primary tumor site
Colon
Rectum
Dwelling
Rural
Urban
ECOG performance status
0
1
2
3
Chemotherapy regimen
5-Fluorouracil, Oxaliplatin, Folinic acid (FOLFOX)
5-Fluorouracil, Irinotecan, Folinic acid (FOLFIRI)
Clinical T-stage
T2
T3
T4
Clinical N-stage
N0
N1
N2
DPYD
GG
GA
MTHFR
CC
CT
TT
CT + TT
DPYD dihydropyrimidine dehydrogenase, MTHFR methylenetetrahydrofolatereductase
112 (61.9)
36 (22.4)
13 (15.7)
49 (38.1)
56 (34.9)
73 (45.3)
25 (15.5)
7 (4.3)
98 (60.9)
39 (24.2)
24 (14.9)
75 (46.6)
49 (30.4)
37 (23)
69 (42.9)
92 (57.1)
153 (95)
8 (5)
71 (91)
7 (9)
54 (69.2)
17 (21.8)
7 (9)
24 (30.8)
41 (49.4)
23 (27.7)
19 (22.9)
54 (65.1)
29 (34.9)
7 (8.4)
76 (91.6)
41 (49.4)
42 (50.6)
41 (49.4)
42 (50.6)
44 (53)
25 (30.1)
13 (15.7)
1 (1.2)
34 (41)
49 (69)
54 (65.1)
16 (19.3)
13 (15.6)
30 (36.1)
30 (36.1)
23 (27.8)
82 (98.8)
1 (1.2)
58 (69.9)
19 (22.9)
6 (7.2)
25 (30.1)
0.343
0.198
0.512
0.224
0.167
–
0.617
0.318
0.0234
0.023
–
0.917
0.701
0.929
MTHFR C677T polymorphisms significantly associ-
ated with 5-FU related several toxicities such as diarrhea
(adjusted OR = 2.14, 95% CI 1.01–4.56, P = 0.048), neutro-
penia (adjusted OR 3.03, 95% CI 1.39–6.60, P = 0.005), and
mucositis (adjusted OR 8.17, 95% CI 1.25–53.61, P = 0.029)
when C/T + T/T compared to C/C genotype.
Impact of MTHFR gene polymorphism on response
Age, sex, performance status, stage, and tumor grade
except BMI have no statistically significant effect on the
response of 5-FU-based chemotherapy (Table 4).
1 3�
Cancer Chemotherapy and Pharmacology
Table 3 Different grades of toxicities if various types caused by 5-FU-based chemotherapy in the patients with different genotypes of DPYD*2A
and MTHFR C677T polymorphism
Gene
Toxicity
Adjusted odds ratio (95% CI)
P value
Genotype
Grade ≤ 2
Anemia
Neutropenia
DPYD*2A
MTHFR
C677T
DPYD*2A
MTHFR
C677T
Thrombocytopenia
DPYD*2A
Leucopenia
Nausea
Vomiting
Diarrhea
Mucositis
MTHFR
C677T
DPYD*2A
MTHFR
C677T
DPYD*2A
MTHFR
C677T
DPYD*2A
MTHFR
C677T
DPYD*2A
MTHFR
C677T
DPYD*2A
MTHFR
C677T
GG (153)
GA (8)
CC (112)
CT (36)
TT (13)
CT (36) + TT (13)
GG (153)
GA (8)
CC (112)
CT (36)
TT (13)
CT (36) + TT (13)
GG (153)
GA (8)
CC (112)
CT (36)
TT (13)
CT (36) + TT (13)
GG (153)
GA (8)
CC (112)
CT (36)
TT (13)
CT (36) + TT (13)
GG (153)
GA (8)
CC (112)
CT (36)
TT (13)
CT (36) + TT (13)
GG (153)
GA (8)
CC (112)
CT (36)
TT (13)
CT (36) + TT (13)
GG (153)
GA (8)
CC (112)
CT (36)
TT (13)
CT (36) + TT (13)
GG (153)
GA (8)
CC (112)
CT (36)
TT (13)
CT (36) + TT (13)
Grade
3 + Grade 4
31
4
20
11
4
15
40
5
25
14
6
20
11
2
11
2
1
3
15
1
9
5
2
7
18
3
14
3
2
5
22
3
17
7
2
9
42
5
29
13
5
20
8
1
6
2
1
3
122
4
92
25
9
34
113
3
87
22
7
29
143
6
101
34
12
46
138
7
103
31
11
42
135
5
98
33
11
44
131
5
95
29
11
40
111
3
83
23
8
29
145
7
106
34
12
46
Reference
4.70 (1.06–20.96)
Reference
1.95 (0.76–5.00)
1.84 (0.45–7.47)
1.93 (0.85–4.37)
Reference
6.47 (1.37–30.51)
Reference
3.03 (1.25–7.30)
4.44 (1.08–18.30)
3.03 (1.39–6.60)
Reference
8.08 (1.00–65.15)
Reference
0.45 (0.08–2.54)
1.16 (0.08–17.24)
0.59 (0.14–2.53)
Reference
1.46 (0.13–16.35)
Reference
2.05 (0.55–7.70)
1.18 (0.10–14.30)
1.51 (0.47–4.84)
Reference
10.06 (1.65–61.26)
Reference
0.73 (0.19–2.75)
0.78 (0.10–6.20)
0.68 (0.22–2.14)
Reference
4.24 (0.80–22.40)
Reference
2.38 (0.82–6.87)
0.54 (0.06–4.98)
1.66 (0.64–4.30)
Reference
5.76 (1.24–26.77)
Reference
2.07 (0.88–4.89)
2.62 (0.72–9.59)
2.14 (1.01–4.56)
Reference
6.09 (0.38–97.46)
Reference
5.73 (0.80–41.27)
7.07 (0.23–217.20)
8.17 (1.25–53.61)
0.042
0.165
0.395
0.118
0.018
0.014
0.039
0.005
0.050
0.364
0.916
0.590
0.759
0.285
0.899
0.490
0.012
0.639
0.811
0.510
0.089
0.109
0.588
0.299
0.026
0.096
0.146
0.048
0.202
0.083
0.263
0.029
1 3�Cancer Chemotherapy and Pharmacology
Table 3 (continued)
Toxicity
Gene
Genotype
Dermatological toxicities
DPYD*2A
MTHFR
C677T
Neurological toxicities
DPYD*2A
MTHFR
C677T
GG (153)
GA (8)
CC (112)
CT (36)
TT (13)
CT (36) + TT (13)
GG (153)
GA (8)
CC (112)
CT (36)
TT (13)
CT (36) + TT (13)
Grade
3 + Grade 4
12
1
8
4
1
5
2
0
2
0
0
0
Grade ≤ 2
Adjusted odds ratio (95% CI)
P value
148
7
104
32
12
44
151
8
110
36
13
49
Reference
1.95 (0.16–23.96)
Reference
0.64 (0.12–3.36)
< 0
0.40 (0.08–2.15)
Reference
–
Reference
–
–
–
0.602
0.594
0.998
0.288
–
–
–
–
–
The highest response was complete response (CR) in 35
patients, partial response (PR) in 46 patients, stable disease
(SD) in 42 patients, and progressive disease (PD) in 16
patients, accounting for a total of 58.27% clinical responses
(C/R + P/R). About 51% patients of C/C, 71.9% patients of
C/T, 81.8% patients of T/T, and 74.4% patients of C/T + T/T
genotypes responded well. Thus, MTHFR C677T genotype
is significantly associated with an increased efficacy of
5-FU (adjusted OR 3.80, 95% CI 1.45–9.92, P = 0.006 when
C/T + T/T compared to C/C genotype) (Table 5).
Discussion
Polymorphisms in the enzymes, which are involved in phar-
macokinetics and pharmacodynamics of a drug, can easily
modulate drug effects and treatment outcome [32]. The anti-
cancer drugs have a narrow therapeutic index [33]. Higher
plasma drug concentrations can cause toxicities, whereas
lower concentrations can lead to reduced efficacy. Therefore,
an optimum plasma drug concentration is important for the
best clinical outcome. Thus, pharmacogenetics can help to
predict the clinical outcome of antitumor therapy based on
individual’s genetic makeup [32]. In this study, we exam-
ined 161 Bangladeshi colorectal cancer patients, receiving
5-fluorouracil-based combined chemotherapy, to correlate
DPYD and MTHFR gene polymorphisms and toxicity as
well as the response to this chemotherapy.
5-FU is biotransformed by DPYD enzyme, and thus, pol-
ymorphism in this DPYD gene can decrease enzyme’s cata-
bolic activity, causing accumulation of 5-FU in blood, and
increasing incidence of toxicities [14, 21]. 5-FU acts as an
anticancer agent mostly by inhibiting TS, which finally sup-
presses DNA synthesis and polymorphism in MTHFR gene
may facilitate this TS inhibition process, increasing 5-FU
response to tumor cells [24–27, 34]. However, although for
Bangladeshi colorectal cancer patients, 5-FU is prescribed
as central to the combined chemotherapy; no clinical stud-
ies have ever been conducted to establish the relationship
between DPYD and MTHFR gene polymorphisms and clini-
cal outcome of fluoropyrimidines for Bangladeshi cancer
patients.
About 30 single nucleotide polymorphisms (SNPs), inser-
tions, and deletions have been identified in DPYD gene [35].
Among them, one splice site mutation at intron 14 (IVS
14+1G>A or *2A, rs3918290) causes the skipping of exon
14 during splicing [36] and produces a nonfunctional protein
[37, 38]. As DPYD enzyme converted 5-FU into 5-dihy-
drofluorouracil (FDHU) which is further metabolized to be
excreted to the urine [17], the decrease in DPYD activity
can accumulate 5-FU in blood and cause toxicities. Cicco-
lini et al. described that 71% patients of total 80 toxic cases
had reduced DPYD activity, and in these patients’ drug,
exposures were 15 times higher than the range observed in
the non-toxic population [39]. Van et al. demonstrated that
43% of the patients having reduced DPYD activity carried
DPYD*2A polymorphism and this polymorphism was the
most abundant one that can cause a decrease in DPYD activ-
ity [40].
In our study, we found that of total 161 colorectal cancer
patients who were treated with 5-FU-based chemotherapy,
78 patients developed grade 3 or grade 4 toxicities and out of
8 patients carrying a variant genotype (G/A), and 7 patients
suffered from grade 3 or grade 4 toxicities. About 9% (n = 7)
of total toxicity cases (n = 78) carry this DPYD*2A poly-
morphism (G/A genotype), although about 87.5% (n = 7) of
the patients carrying this polymorphism (n = 8) were suffer-
ing from grade 3 or grade 4 toxicities. We found a signifi-
cant association between A allele of DPYD*2A and grade
3 or grade 4 anemia (P = 0.042), neutropenia (P = 0.018),
1 3�
Cancer Chemotherapy and Pharmacology
Table 4 Correlations between clinicopathological features and response status in 139 colorectal cancer patients
Characteristics of the patients
Responders (n = 81)
Total cases (n = 139)
Non-responders
(n = 58)
P value
14 (10.1)
125 (89.9)
63 (45.3)
47 (33.8)
29 (20.9)
83 (59.7)
56 (40.3)
78 (56.1)
61 (43.9)
66 (47.5)
73 (52.5)
Age (years)
< 45
45–60
> 60
Sex
Male
Female
Body Mass Index (BMI)
< 18.5
18.5–30
Primary tumor site
Colon
Rectum
Dwelling
Rural
Urban
ECOG performance status
0
1
2
3
Chemotherapy regimen
5-Fluorouracil, Oxaliplatin, Folinic acid (FOLFOX)
5-Fluorouracil, Irinotecan, Folinic acid (FOLFIRI)
Clinical T-stage
T2
T3
T4
Clinical N-stage
N0
N1
N2
DPYD dihydropyrimidine dehydrogenase, MTHFR methylenetetrahydrofolatereductase
50 (36)
63 (45.3)
21 (15.1)
5 (3.6)
66 (47.5)
44 (31.7)
29 (20.8)
60 (43.1)
79 (56.8)
87 (62.6)
35 (25.2)
17 (12.2)
40 (49.3)
25 (30.9)
16 (19.8)
48 (59.3)
33 (40.7)
1 (1.2)
80 (98.8)
41 (50.6)
40 (49.4)
35 (43.2)
46 (56.7)
49 (60.5)
22 (27.2)
10 (12.3)
0 (0)
40 (49.4)
41 (50.6)
51 (63)
19 (23.4)
11 (13.6)
40 (49.4)
25 (30.9)
16 (19.7)
Table 5 Comparison of
responders and non-responders
with MTHFR 677C>T
polymorphism
MTHFR 677C>T (rs1801133) (n = 139)
Genotype
23 (39.7)
22 (37.9)
13 (22.4)
35 (60.3)
23 (29.7)
13 (22.4)
45 (77.6)
37 (63.8)
21 (36.2)
31 (53.4)
27 (46.6)
1 (1.7)
41 (70.7)
11 (19)
5 (8.6)
20 (34.5)
38 (65.5)
36 (62.1)
16 (27.5)
6 (10.4)
26 (44.8)
19 (32.8)
13 (22.4)
0.516
0.888
0.0001
0.123
0.233
–
0.08
0.771
0.861
Adjusted OR (95% CI)
P value
Responders
(CR + PR) (n = 81)
49 (60.5)
23 (28.4)
9 (11.1)
32 (39.5)
Non-responders
(SD + PD) (n = 58)
47 (81)
9 (15.5)
2 (3.5)
11 (19)
Reference
CC (96)
3.57 (1.20–10.66)
CT (32)
4.19 (0.79–22.19)
TT (11)
3.80 (1.45–9.92)
CT (32) + TT (11)
CR complete response, PR partial response, SD stable disease, PD progressive disease
0.023
0.092
0.006
thrombocytopenia (P = 0.050), nausea (P = 0.012), and diar-
rhea (P = 0.026) when G/A genotype compared to G/G geno-
type. This result is not conflicting with other clinical studies
[41, 42]. DPYD*2A polymorphism was found in 50% of
cancer patients with the fourth stage of neutropenia [43].
Van et al. [44] studied on 95 patients and 60 of them suffered
1 3�Cancer Chemotherapy and Pharmacology
from grade 3 or 4 toxicities, and of these 60 patients, 28%
cases carried this DPYD*2A polymorphism. Schwab et al.
[45] demonstrated that the sensitivity of DPYD*2A geno-
typing for overall toxicity was 5.5% which was significantly
associated with 5-FU related toxicities. Due to low allele
frequency, Gross et al. [46] did not get any significant rela-
tionship as only five cases with this polymorphism, out of 92
toxic cases were found in this study. Several studies found
some other polymorphisms c.496A>G, c.1679T>G, and
c.2846A>T significant for decreased DPYD activity. But
we did not include these polymorphisms in our study, as
we thought these would create limited impact in our study,
owing to a low allele frequency of these gene variants in
this studied population. DPYD c.496A>G polymorphism
is infrequent in Korean [47] and Japanese [48] populations.
DPYD c.1679T>G and c.2846A>T polymorphisms are very
rare and thus account for only a very small part of the 5-FU
toxicities [41, 45, 46, 49]. According to the 1000 genome
project DPYD, c.1679T>G and c.2846A>T polymorphisms
are absent in Bangladesh and DPYD c.496A>G polymor-
phism is extremely rare in Bangladeshi population.
We observed that only 5% patients carry G/A genotype,
but about 87.5% of these patients with G/A genotype suf-
fered from grade 3 or grade 4.
In our study, we found only eight patients with DPYD*2A
polymorphism with allele frequency of 2.48%, all of them
were in heterozygote states, and no mutant homozygote was
found. Although this SNP can cause a significant decrease
in DPYD activity and subsequent increase in 5-FU related
toxicities, this polymorphism frequency is not very much
high in our population. In other Asian countries, this poly-
morphism is also not very much frequent. No mutation was
found in normal Japanese people and heterozygotes were
found in Taiwanese people with an allelic frequency of 0.027
[38].
Methylenetetrahydrofolatereductase (MTHFR) is another
gene that was examined in this study. MTHFR enzyme cat-
alyzes the conversion of 5, 10-methylene-THF (CH2THF)
to 5-methyltetrahydrofolate (CH3THF) [27]. Frosst et al.
described that C677T polymorphism can cause a 30%
reduction in enzymatic activity. The presence of 677T vari-
ant can cause accumulation of CH2THF in cells, which can
modulate the efficacy of 5-FU [50]. Cohen et al. declared
this C677T polymorphism as a genomic predictor of clini-
cal response to fluoropyrimidine-based chemotherapy [24].
This is because of 5-FdUMP, a metabolite of 5-FU irrevers-
ibly forms a ternary complex with TS and CH2THF [25].
This complex inhibits the activity of TS, which leads to a
decrease in concentration of dTMP and then finally inhi-
bition of DNA synthesis. Sohn et al. proved this in both
in vitro and in vivo studies, where he showed that T allele
increases chemosensitivity in colon cancer cells [25]. Again,
an increased in response can be predicted to cause toxicity
higher in patients carrying 677T allele. There are some other
polymorphisms present in MTHFR gene, but these are not
very much significant. The 1298A>C transition also leads to
decreased enzyme activity, although not to the same extent
as the 677T allele [51]. Etienne demonstrated that response
rate was not related to 1298A>C genotype, but was signifi-
cantly linked to 677C>T genotype [52]. This influenced us
to include MTHFR C677T polymorphism only in this study
to assess the impact of this polymorphism in the outcome
of 5-FU treatment.
In our study, we found that MTHFR C677T polymor-
phism is significantly associated with increased tumor
response to 5-FU (P = 0.006) and we also found that this
can increase the risk of developing grade 3 or 4 neutrope-
nia, diarrhea, and mucositis. In toxicity assessment, we got
a significant relationship between 677T allele and 5-FU-
induced neutropenia, diarrhea, and mucositis. In other types
of toxicities such as anemia, thrombocytopenia, leukopenia,
vomiting, nausea, dermatological toxicities, and neurologi-
cal toxicities, we did not find any significant relationship.
Several studies found a significant relationship between
MTHFR C677T polymorphism with increased tumor
response to 5-FU [24, 53, 54]. Etienne et al. also demon-
strated that MTHFR C677T genotype was linked to clinical
response (P = 0.042), with the rare allele linked to improved
response [55]. Several population studies revealed contradic-
tory and inconsistent conclusions. Marcuello et al. could
not confirm this association [56], whereas Chua et al. [33]
did not find any significant relationship between MTHFR
C677T polymorphism and improved FOLFOX efficacy.
In our study, MTHFR 677T allele is significantly linked
to grade 3 or 4 diarrhea only. Chua et al. [33] found that
patients with the T/T genotype suffered a significantly higher
incidence of grades 3–4 diarrhea. Etienne et al. [55] found
no association with toxicity. Sharma et al. [57] reported that
patients with the MTHFR 677T/T genotype had a lower inci-
dence of grade 2 or 3 toxicity than patients with C/T and
C/C genotypes.
Different studies showed different types of association
between MTHFR C677T polymorphism and efficacy and
toxicity of 5-FU. However, it could be noted that chemo-
therapy regimen and cancer types were not same in all the
cases. This could explain these differences in different clini-
cal studies. However, our study shows a clear increase in
tumor response to 5-FU and incidence of diarrhea when
given to colorectal cancer patients.
DPYD*2A polymorphism that skips exon 14 during
splicing is an obvious predictor for decreasing the DPYD
enzymatic activity. Thus, patients having this mutation can
develop severe life-threatening toxicities when they are
treated with 5-FU-based chemotherapy. Thus, this polymor-
phism should be taken as a pharmacogenetic determinant for
5-FU-related toxicities. Our observations on the association
1 3�
Cancer Chemotherapy and Pharmacology
of MTHFR C677T for response and toxicity require con-
firmation in large studies that should be conducted in the
same chemotherapy regimen and in the same types of cancer
patients.
Acknowledgements We are very grateful to all the patients, controls,
physicians, and nurses. We are also grateful to the Department of Clini-
cal Pharmacy and Pharmacology for the partial support to conduct
this research project. The authors have no other relevant affiliations or
financial involvement with any organization.
Compliance with ethical standards
Conflict of interest The authors declare that there are no conflicts of
interest.
Ethical approval All procedures performed in this study involving
human participants were in accordance with the ethical standards of the
institutional research committee and with the 1964 Helsinki declaration
and its later amendments or comparable ethical standards.
Informed consent Informed consent was obtained from all individual
participants included in the study.
References
1. Ferlay J, Ferlay J, Soerjomataram I et al. (2013) GLOBOCAN
2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC
CancerBase No. 11 [Internet]. Lyon, France: International Agency
for Research on Cancer. http://globocan.iarc.fr. Accessed on 10
Apr 2015
2. Heidelberger C, Chaudhuri NK, Danneberg P et al (1957) Fluori-
nated pyrimidines, a new class of tumour-inhibitory compounds.
Nature 179(4561):663–666
3. Giacchetti S, Perpoint B, Zidani R et al (2000) Phase III multi-
center randomized trial of oxaliplatin added to chronomodulated
fluorouracil–leucovorin as first-line treatment of metastatic colo-
rectal cancer. J Clin Oncol 18(1):136–147
4. DeGramont A, Figer A, Seymour M et al (2000) Leucovorin and
fluorouracil with or without oxaliplatin as first-line treatment in
advanced colorectal cancer. J Clin Oncol 18(16):2938–2947
5. Hurwitz H, Fehrenbacher L, Novotny W et al (2004) Bevacizumab
plus irinotecan, fluorouracil, and leucovorin for metastatic colo-
rectal cancer. N Engl J Med 350(23):2335–2342
6. Ezzeldin H, Diasio R (2004) Dihydropyrimidine dehydrogenase
deficiency, a pharmacogenetic syndrome associated with poten-
tially life-threatening toxicity following 5-fluorouracil administra-
tion. Clin Colorectal Cancer 4(3):181–189
7. Colucci G, Gebbia V, Paoletti G et al (2005) Phase III randomized
trial of FOLFIRI versus FOLFOX4 in the treatment of advanced
colorectal cancer: a multicenter study of the Gruppo Oncologico
Dell’ Italia Meridionale. J Clin Oncol 23(22):4866–4875
8. Chung KY, Saltz LB (2007) Adjuvant therapy of colon cancer:
current status and future directions. Cancer J 13(3):192–197
9. Board RE, Valle JW (2007) Metastatic colorectal cancer: current
systemic treatment options. Drugs 67(13):1851–1867
10. Sauer R, Liersch T, Merkel S et al (2012) Preoperative versus
postoperative chemoradiotherapy for locally advanced rectal
cancer: results of the German CAO/ARO/AIO-94 randomized
phase III trial after a median follow-up of 11 years. J Clin Oncol
30(16):1926–1933
11. Asmis T, Berry S, Cosby R et al (2014) Cancer Care Ontario’s
Gastrointestinal Disease Site Group. Strategies of sequential
therapies in unresectable metastatic colorectal cancer: a meta-
analysis. Curr Oncol 21(6):318–328
12. Chua W, Kho PS, Moore MM et al (2011) Clinical, labora-
tory and molecular factors predicting chemotherapy efficacy
and toxicity in colorectal cancer. Crit Rev Oncol Hematol
79(3):224–250
13. Hofheinz RD, Wenz F, Post S at al (2012) Chemoradiotherapy
with capecitabine versus fluorouracil for locally advanced rectal
cancer: a randomised, multicentre, non-inferiority, phase 3 trial.
Lancet Oncol 13(6):579–588
14. Li W, Xu J, Shen L, Liu T et al (2014) Phase II study of weekly
irinotecan and capecitabine treatment in metastatic colorectal
cancer patients. BMC Cancer 14:986
15. Meta-Analysis Group In Cancer, Lévy E, Piedbois P, Buyse M
et al (1998) Toxicity of fluorouracil in patients with advanced
colorectal cancer: effect of administration schedule and prog-
nostic factors. J Clin Oncol 16 (11):3537–3541
16. Diasio RB, Harris BE (1989) Clinical pharmacology of 5-fluo-
rouracil. Clin Pharmacokinet 16(4):215–237
17. Mattison LK, Soong R, Diasio RB (2002) Implications of dihy-
dropyrimidine dehydrogenase on 5-fluorouracil pharmacogenet-
ics and pharmacogenomics. Pharmacogenomics 3(4):485–492
18. Thorn CF, Marsh S, Carrillo MW et al (2011) PharmGKB sum-
mary: fluoropyrimidine pathways. Pharmacogenet Genomics
21(4):237–242
19. Van Kuilenburg AB, Meinsma R, Zonnenberg BA et al (2003)
Dihydropyrimidinase deficiency and severe 5-fluorouracil toxic-
ity. Clin Cancer Res 9(12):4363–4367
20. Van Kuilenburg AB, Maring JG, Schalhorn A et al. (2008) Phar-
macokinetics of 5-fluorouracil in patients heterozygous for the
IVS14 + 1G > A mutation in the dihydropyrimidine dehydro-
genase gene. Nucl Nucl Nucl Acids 27(6):692–698
21. Amstutz U, Froehlich TK, Largiadèr CR (2011) Dihydropy-
rimidine dehydrogenase gene as a major predictor of severe
5-fluorouracil toxicity. Pharmacogenomics 12(9):1321–1336
22. Kaldate RR, Haregewoin A, Grier CE et al (2012) Modeling
the 5-fluorouracil area under the curve versus dose relationship
to develop a pharmacokinetic dosing algorithm for colorectal
cancer patients receiving FOLFOX6. Oncologist 17(3):296–302
23. Etienne MC, Lagrange JL, Dassonville O et al (1994) Popu-
lation study of dihydropyrimidine dehydrogenase in cancer
patients. J Clin Oncol 12(11):2248–2253
24. Cohen V, Panet-Raymond V, Sabbaghian N, Morin I et al (2003)
Methylenetetrahydrofolatereductase polymorphism in advanced
colorectal cancer: a novel genomic predictor of clinical response
to fluoropyrimidine-based chemotherapy. Clin Cancer Res
9(5):1611–1615
25. Sohn KJ, Croxford R, Yates Z et al (2004) Effect of the methyl-
enetetrahydrofolatereductase C677T polymorphism on chemo-
sensitivity of colon and breast cancer cells to 5-fluorouracil and
methotrexate. J Natl Cancer Inst 96:134–144
26. Toffoli G, De Mattia E (2008) Pharmacogenetic relevance of
MTHFR polymorphisms. Pharmacogenomics 9(9):1195–1206
27. Kantar M, Kosova B, Cetingul N et al (2009) Methylenetet-
rahydrofolatereductase C677T and A1298C gene polymor-
phisms and therapy-related toxicity in children treated for acute
lymphoblastic leukemia and non-Hodgkin lymphoma. Leuk
Lymphoma 50(6):912–917
28. World Medical Association Declaration of Helsinki (2008)
Ethical principles for medical research involving human sub-
jects. Adopted by the 18th WMA General Assembly, Helsinki,
Finland, June 1964, and amended by the 59th WMA General
Assembly Seoul, South Korea
1 3�Cancer Chemotherapy and Pharmacology
29. National Cancer Institute Common Terminology Criteria for
Adverse Events, Version 3.0. http://ctep.cancer.gov/protocolD-
evelopment/electronic_applications/docs/ctcaev3.pdf
30. Response Evaluation Criteria in Solid Tumors (RECST) https://
www.eortc.be/Recist/documents/ RECISTGuidelines.pdf
31. Islam MS, Ahmed MU, Sayeed MS et al (2013) Lung cancer
risk in relation to nicotinic acetylcholine receptor, CYP2A6 and
CYP1A1 genotypes in the Bangladeshi population. Clin Chim
Acta 416:11–19
32. Ma Q, Lu AY (2011) Pharmacogenetics, pharmacogenomics, and
individualized medicine. Pharmacol Rev 63(2):437–459
33. Stewart CF, Schuetz EG (2000) Need and potential for predictive
tests of hepatic metabolism of anticancer drugs. Clin Cancer Res
6(9):3391–3392
34. Chua W, Goldstein D, Lee CK et al (2009) Molecular markers of
response and toxicity to FOLFOX chemotherapy in metastatic
colorectal cancer. Br J Cancer 101(6):998–1004
35. Huang RS, Ratain MJ (2009) Pharmacogenetics and pharmacog-
enomics of anticancer agents. CA Cancer J Clin 59(1):42–55
36. Van Kuilenburg AB, Vreken P, Beex LV et al (1997) Heterozygo-
sity for a point mutation in an invariant splice donor site of dihy-
dropyrimidine dehydrogenase and severe 5-fluorouracil related
toxicity. Eur J Cancer 33(13):2258–2264
37. Vreken P, Vankuilenburg ABP, Meinsma R et al (1996) A point
mutation in an invariant splice donor site leads to exon skipping
in two unrelated Dutch patients with dihydropyrimidine dehydro-
genase deficiency. J Inherit Metab Dis 19(5):645–654
38. Wei X, McLeod HL, McMurrough J et al (1996) Molecular basis
of the human dihydropyrimidine dehydrogenase deficiency and
5-fluorouracil toxicity. J Clin Invest 98:610–615
39. Ciccolini J, Mercier C, Evrard A et al (2006) A rapid and inex-
pensive method for anticipating severe toxicity to fluorouracil and
fluorouracil-based chemotherapy. Ther Drug Monit 8(5):678–685
40. Van Kuilenburg AB, Haasjes J, Meinsma R et al (2000) Dihydro-
pyrimidine dehydrogenase (DPD) deficiency: novel mutations in
the DPD gene. Adv Exp Med Biol 486:247–250
41. Boisdron-Celle M, Remaud G, Traore S et al (2007) 5-fluoroura-
cil-related severe toxicity: a comparison of different methods for
the pretherapeutic detection of dihydropyrimidine dehydrogenase
deficiency. Cancer Lett 249(2):271–282
42. Terrazzino S, Cargnin S, Del Re M et al (2013) DPYD IVS14 +
1G > A and 2846A > T genotyping for the prediction of severe
fluoropyrimidine-related toxicity: a meta-analysis. Pharmacog-
enomics 14(11):1255–1272
43. Van Kuilenburg AB, Haasjes J, Van Lenthe H et al (2000) Dihy-
dropyrimidine dehydrogenase deficiency and 5-fluorouracil asso-
ciated toxicity. Adv Exp Med Biol 486:251–255
44. Van Kuilenburg AB, Dobritzsch D, Meinsma R et al (2002) Novel
disease-causing mutations in the dihydropyrimidine dehydroge-
nase gene interpreted by analysis of the three dimensional protein
structures. Biochem J 364(Pt 1):157–163
45. Schwab M, Zanger UM, Marx C et al (2008) Role of genetic and
nongenetic factors for fluorouracil treatment-related severe toxic-
ity: a prospective clinical trial by the German 5-FU Toxicity Study
Group. J Clin Oncol 26(13):2131–2138
46. Gross E, Busse B, Riemenschneider M et al. (2008) Strong asso-
ciation of a common dihydropyrimidine dehydrogenase gene
polymorphism with fluoropyrimidine-related toxicity in cancer
patients. PLoS One 3 (12), e4003
47. Cho H, Park Y, Kang W, Kim J, Lee S (2007) Thymidylate syn-
thase (TYMS) and dihydropyrimidine dehydrogenase (DPYD)
polymorphisms in the Korean population for prediction of 5-fluo-
rouracil-associated toxicity. Ther Drug Monit 29(2):190–196
48. Maekawa K, Saeki M, Saito Y et al (2007) Genetic variations and
haplotype structures of the DPYD gene encoding dihydropyrimi-
dine dehydrogenase in Japanese and their ethnic differences. J
Hum Genet 52(10):804–819
49. Morel A, Boisdron-Celle M, Fey L et al (2006) Clinical relevance
of different dihydropyrimidine dehydrogenase gene single nucleo-
tide polymorphisms on 5-fluorouracil tolerance. Mol Cancer Ther
5(11):2895–2904
50. Frosst P, Blom HJ, Milos R et al (1995) A candidate genetic risk
factor for vascular disease: a common mutation in methylenetet-
rahydrofolatereductase. Nat Genet 10:111–113
51. Yeh CC, Lai CY, Chang SN et al (2017) Polymorphisms of
MTHFR C677T and A1298C associated with survival in patients
with colorectal cancer treated with 5-fluorouracil-based chemo-
therapy. Int J Clin Oncol 2:1–10
52. Etienne MC, Formento JL, Chazal M et al (2004) Methylenetet-
rahydrofolate reductase gene polymorphisms and response to fluo-
rouracil-based treatment in advanced colorectal cancer patients.
Pharmacogenet Genom 14(12):785–792
53. Etienne MC, Ilc K, Formento JL et al (2004) Thymidylate syn-
thase and methylenetetrahydrofolatereductase gene polymor-
phisms: relationships with 5-fluorouracil sensitivity. Br J Cancer
90:526–534
54. Jakobsen A, Nielsen JN, Gyldenkerne N, Lindeberg J (2005) Thy-
midylate synthase and methylenetetrahydrofolatereductase gene
polymorphism in normal tissue as predictors of fluorouracil sen-
sitivity. J Clin Oncol 23:1365–1369
55. Etienne-Grimaldi MC, Milano G, Maindrault-Goebel F et al
(2010) Methylenetetrahydrofolatereductase (MTHFR) gene poly-
morphisms and FOLFOX response in colorectal cancer patients.
Br J Clin Pharmacol 69(1):58–66
56. Marcuello E, Altés A, Menoyo A et al (2006) Methylenetetrahy-
drofolatereductase gene polymorphisms: genomic predictors of
clinical response to fluoropyrimidine-based chemotherapy? Can-
cer Chemother Pharmacol 57:835–840
57. Sharma R, Hoskins JM, Rivory LP et al (2008) Thymidylate
synthase and methylenetetrahydrofolatereductase gene polymor-
phisms and toxicity to capecitabine in advanced colorectal cancer
patients. Clin Cancer Res 14:817–825
1 3�
#### replace unknown text
text = text.replace("\xa0","").replace("\n"," ")
### import regex module
import re
variant_compile = re.compile(r"(rs)\d+")
gene_compile = re.compile(r"[A-Z]+\d+\w+")
poly_compile = re.compile(r"[A-Z]{2,9}\*?\w+")
for pos in variant_compile.finditer(text):
print(pos)
<re.Match object; span=(10264, 10273), match='rs3918290'>
<re.Match object; span=(10293, 10302), match='rs1801133'>
<re.Match object; span=(11935, 11944), match='rs3918290'>
<re.Match object; span=(12096, 12105), match='rs1801133'>
<re.Match object; span=(23570, 23579), match='rs3918290'>
<re.Match object; span=(26238, 26247), match='rs1801133'>
for gene in gene_compile.finditer(text):
print(gene)
<re.Match object; span=(116, 121), match='C677T'>
<re.Match object; span=(944, 949), match='C677T'>
<re.Match object; span=(1517, 1522), match='C677T'>
<re.Match object; span=(1741, 1746), match='C677T'>
<re.Match object; span=(5071, 5076), match='C677T'>
<re.Match object; span=(10286, 10291), match='C677T'>
<re.Match object; span=(10458, 10463), match='CH4IV'>
<re.Match object; span=(10503, 10508), match='C677T'>
<re.Match object; span=(11923, 11928), match='IVS14'>
<re.Match object; span=(12089, 12094), match='C677T'>
<re.Match object; span=(14478, 14483), match='C677T'>
<re.Match object; span=(17866, 17871), match='C677T'>
<re.Match object; span=(18595, 18600), match='C677T'>
<re.Match object; span=(18725, 18730), match='C677T'>
<re.Match object; span=(18747, 18752), match='C677T'>
<re.Match object; span=(18838, 18843), match='C677T'>
<re.Match object; span=(18860, 18865), match='C677T'>
<re.Match object; span=(18882, 18887), match='C677T'>
<re.Match object; span=(18904, 18909), match='C677T'>
<re.Match object; span=(18926, 18931), match='C677T'>
<re.Match object; span=(18948, 18953), match='C677T'>
<re.Match object; span=(20770, 20775), match='C677T'>
<re.Match object; span=(20817, 20822), match='C677T'>
<re.Match object; span=(21616, 21621), match='C677T'>
<re.Match object; span=(29447, 29453), match='CH2THF'>
<re.Match object; span=(29486, 29492), match='CH3THF'>
<re.Match object; span=(29530, 29535), match='C677T'>
<re.Match object; span=(29657, 29663), match='CH2THF'>
<re.Match object; span=(29748, 29753), match='C677T'>
<re.Match object; span=(29960, 29966), match='CH2THF'>
<re.Match object; span=(30768, 30773), match='C677T'>
<re.Match object; span=(30919, 30924), match='C677T'>
<re.Match object; span=(31537, 31542), match='C677T'>
<re.Match object; span=(31658, 31663), match='C677T'>
<re.Match object; span=(31997, 32002), match='C677T'>
<re.Match object; span=(32554, 32559), match='C677T'>
<re.Match object; span=(33383, 33388), match='C677T'>
<re.Match object; span=(38264, 38269), match='IVS14'>
<re.Match object; span=(39332, 39337), match='C677T'>
<re.Match object; span=(39679, 39684), match='C677T'>
<re.Match object; span=(39689, 39695), match='A1298C'>
<re.Match object; span=(40588, 40594), match='CYP2A6'>
<re.Match object; span=(40600, 40606), match='CYP1A1'>
<re.Match object; span=(42592, 42597), match='IVS14'>
<re.Match object; span=(44571, 44576), match='C677T'>
<re.Match object; span=(44581, 44587), match='A1298C'>
for poly in poly_compile.finditer(text):
print(poly)
<re.Match object; span=(81, 89), match='ORIGINAL'>
<re.Match object; span=(90, 97), match='ARTICLE'>
<re.Match object; span=(99, 106), match='DPYD*2A'>
<re.Match object; span=(110, 115), match='MTHFR'>
<re.Match object; span=(751, 755), match='ADRs'>
<re.Match object; span=(926, 933), match='DPYD*2A'>
<re.Match object; span=(938, 943), match='MTHFR'>
<re.Match object; span=(1150, 1154), match='DPYD'>
<re.Match object; span=(1160, 1165), match='MTHFR'>
<re.Match object; span=(1385, 1392), match='DPYD*2A'>
<re.Match object; span=(1511, 1516), match='MTHFR'>
<re.Match object; span=(1723, 1730), match='DPYD*2A'>
<re.Match object; span=(1735, 1740), match='MTHFR'>
<re.Match object; span=(1892, 1896), match='DPYD'>
<re.Match object; span=(1898, 1903), match='MTHFR'>
<re.Match object; span=(1958, 1961), match='CRC'>
<re.Match object; span=(2211, 2214), match='CRC'>
<re.Match object; span=(2706, 2709), match='CRC'>
<re.Match object; span=(3229, 3232), match='CRC'>
<re.Match object; span=(3775, 3779), match='DPYD'>
<re.Match object; span=(3912, 3916), match='FDHU'>
<re.Match object; span=(3925, 3929), match='FDHU'>
<re.Match object; span=(4233, 4236), match='SNP'>
<re.Match object; span=(4276, 4280), match='DPYD'>
<re.Match object; span=(4379, 4383), match='DPYD'>
<re.Match object; span=(4507, 4512), match='MTHFR'>
<re.Match object; span=(4773, 4776), match='UMP'>
<re.Match object; span=(4860, 4863), match='THF'>
<re.Match object; span=(4927, 4930), match='DNA'>
<re.Match object; span=(4952, 4957), match='MTHFR'>
<re.Match object; span=(5003, 5006), match='THF'>
<re.Match object; span=(5046, 5049), match='THF'>
<re.Match object; span=(5065, 5070), match='MTHFR'>
<re.Match object; span=(5128, 5133), match='MTHFR'>
<re.Match object; span=(5178, 5181), match='THF'>
<re.Match object; span=(5211, 5214), match='THF'>
<re.Match object; span=(5359, 5363), match='DPYD'>
<re.Match object; span=(5369, 5374), match='MTHFR'>
<re.Match object; span=(5573, 5578), match='MTHFR'>
<re.Match object; span=(5911, 5916), match='NICRH'>
<re.Match object; span=(6029, 6034), match='NICRH'>
<re.Match object; span=(6167, 6173), match='FOLFOX'>
<re.Match object; span=(6227, 6234), match='FOLFIRI'>
<re.Match object; span=(6740, 6743), match='AST'>
<re.Match object; span=(7475, 7478), match='CBC'>
<re.Match object; span=(8328, 8331), match='WMA'>
<re.Match object; span=(8815, 8820), match='CTCAE'>
<re.Match object; span=(8845, 8849), match='DPYD'>
<re.Match object; span=(8854, 8859), match='MTHFR'>
<re.Match object; span=(9059, 9065), match='RECIST'>
<re.Match object; span=(9140, 9146), match='RECIST'>
<re.Match object; span=(9776, 9781), match='MTHFR'>
<re.Match object; span=(10033, 10036), match='DNA'>
<re.Match object; span=(10214, 10217), match='DNA'>
<re.Match object; span=(10235, 10242), match='DPYD*2A'>
<re.Match object; span=(10246, 10250), match='DPYD'>
<re.Match object; span=(10280, 10285), match='MTHFR'>
<re.Match object; span=(10339, 10343), match='SNPs'>
<re.Match object; span=(10415, 10418), match='PCR'>
<re.Match object; span=(10419, 10423), match='RFLP'>
<re.Match object; span=(10458, 10463), match='CH4IV'>
<re.Match object; span=(10485, 10492), match='DPYD*2A'>
<re.Match object; span=(10497, 10502), match='MTHFR'>
<re.Match object; span=(10509, 10512), match='PCR'>
<re.Match object; span=(10590, 10593), match='PCR'>
<re.Match object; span=(10654, 10657), match='SNP'>
<re.Match object; span=(10789, 10792), match='SNP'>
<re.Match object; span=(10867, 10870), match='SNP'>
<re.Match object; span=(11064, 11068), match='NCBI'>
<re.Match object; span=(11405, 11408), match='ORs'>
<re.Match object; span=(11440, 11443), match='CIs'>
<re.Match object; span=(11516, 11519), match='BMI'>
<re.Match object; span=(11540, 11544), match='ECOG'>
<re.Match object; span=(11815, 11819), match='SPSS'>
<re.Match object; span=(11885, 11890), match='CHIV4'>
<re.Match object; span=(11918, 11922), match='DPYD'>
<re.Match object; span=(11923, 11928), match='IVS14'>
<re.Match object; span=(12083, 12088), match='MTHFR'>
<re.Match object; span=(12639, 12642), match='BMI'>
<re.Match object; span=(12701, 12707), match='FOLFOX'>
<re.Match object; span=(12723, 12730), match='FOLFIRI'>
<re.Match object; span=(12919, 12922), match='BMI'>
<re.Match object; span=(12991, 12994), match='WHO'>
<re.Match object; span=(13086, 13092), match='FOLFOX'>
<re.Match object; span=(13136, 13143), match='FOLFIRI'>
<re.Match object; span=(14309, 14316), match='DPYD*2A'>
<re.Match object; span=(14437, 14444), match='DPYD*2A'>
<re.Match object; span=(14472, 14477), match='MTHFR'>
<re.Match object; span=(14680, 14684), match='DPYD'>
<re.Match object; span=(14688, 14693), match='MTHFR'>
<re.Match object; span=(14760, 14763), match='BMI'>
<re.Match object; span=(15173, 15180), match='DPYD*2A'>
<re.Match object; span=(16044, 16051), match='DPYD*2A'>
<re.Match object; span=(16862, 16865), match='BMI'>
<re.Match object; span=(16934, 16938), match='ECOG'>
<re.Match object; span=(17030, 17036), match='FOLFOX'>
<re.Match object; span=(17080, 17087), match='FOLFIRI'>
<re.Match object; span=(17141, 17145), match='DPYD'>
<re.Match object; span=(17152, 17157), match='MTHFR'>
<re.Match object; span=(17175, 17179), match='DPYD'>
<re.Match object; span=(17213, 17218), match='MTHFR'>
<re.Match object; span=(17860, 17865), match='MTHFR'>
<re.Match object; span=(18198, 18203), match='MTHFR'>
<re.Match object; span=(18295, 18298), match='BMI'>
<re.Match object; span=(18576, 18583), match='DPYD*2A'>
<re.Match object; span=(18589, 18594), match='MTHFR'>
<re.Match object; span=(18710, 18717), match='DPYD*2A'>
<re.Match object; span=(18719, 18724), match='MTHFR'>
<re.Match object; span=(18732, 18739), match='DPYD*2A'>
<re.Match object; span=(18741, 18746), match='MTHFR'>
<re.Match object; span=(18772, 18779), match='DPYD*2A'>
<re.Match object; span=(18832, 18837), match='MTHFR'>
<re.Match object; span=(18845, 18852), match='DPYD*2A'>
<re.Match object; span=(18854, 18859), match='MTHFR'>
<re.Match object; span=(18867, 18874), match='DPYD*2A'>
<re.Match object; span=(18876, 18881), match='MTHFR'>
<re.Match object; span=(18889, 18896), match='DPYD*2A'>
<re.Match object; span=(18898, 18903), match='MTHFR'>
<re.Match object; span=(18911, 18918), match='DPYD*2A'>
<re.Match object; span=(18920, 18925), match='MTHFR'>
<re.Match object; span=(18933, 18940), match='DPYD*2A'>
<re.Match object; span=(18942, 18947), match='MTHFR'>
<re.Match object; span=(20755, 20762), match='DPYD*2A'>
<re.Match object; span=(20764, 20769), match='MTHFR'>
<re.Match object; span=(20802, 20809), match='DPYD*2A'>
<re.Match object; span=(20811, 20816), match='MTHFR'>
<re.Match object; span=(21610, 21615), match='MTHFR'>
<re.Match object; span=(22517, 22521), match='DPYD'>
<re.Match object; span=(22526, 22531), match='MTHFR'>
<re.Match object; span=(22638, 22642), match='DPYD'>
<re.Match object; span=(22684, 22688), match='DPYD'>
<re.Match object; span=(22913, 22916), match='DNA'>
<re.Match object; span=(22947, 22952), match='MTHFR'>
<re.Match object; span=(23267, 23271), match='DPYD'>
<re.Match object; span=(23276, 23281), match='MTHFR'>
<re.Match object; span=(23422, 23426), match='SNPs'>
<re.Match object; span=(23481, 23485), match='DPYD'>
<re.Match object; span=(23549, 23552), match='IVS'>
<re.Match object; span=(23685, 23689), match='DPYD'>
<re.Match object; span=(23742, 23746), match='FDHU'>
<re.Match object; span=(23828, 23832), match='DPYD'>
<re.Match object; span=(23977, 23981), match='DPYD'>
<re.Match object; span=(24176, 24180), match='DPYD'>
<re.Match object; span=(24199, 24206), match='DPYD*2A'>
<re.Match object; span=(24298, 24302), match='DPYD'>
<re.Match object; span=(24671, 24678), match='DPYD*2A'>
<re.Match object; span=(24901, 24908), match='DPYD*2A'>
<re.Match object; span=(25425, 25428), match='BMI'>
<re.Match object; span=(25497, 25501), match='ECOG'>
<re.Match object; span=(25593, 25599), match='FOLFOX'>
<re.Match object; span=(25643, 25650), match='FOLFIRI'>
<re.Match object; span=(25704, 25708), match='DPYD'>
<re.Match object; span=(25742, 25747), match='MTHFR'>
<re.Match object; span=(26196, 26201), match='MTHFR'>
<re.Match object; span=(26224, 26229), match='MTHFR'>
<re.Match object; span=(27135, 27142), match='DPYD*2A'>
<re.Match object; span=(27424, 27431), match='DPYD*2A'>
<re.Match object; span=(27502, 27509), match='DPYD*2A'>
<re.Match object; span=(27911, 27915), match='DPYD'>
<re.Match object; span=(28132, 28136), match='DPYD'>
<re.Match object; span=(28221, 28225), match='DPYD'>
<re.Match object; span=(28403, 28407), match='DPYD'>
<re.Match object; span=(28477, 28481), match='DPYD'>
<re.Match object; span=(28744, 28751), match='DPYD*2A'>
<re.Match object; span=(28891, 28894), match='SNP'>
<re.Match object; span=(28932, 28936), match='DPYD'>
<re.Match object; span=(29325, 29330), match='MTHFR'>
<re.Match object; span=(29382, 29387), match='MTHFR'>
<re.Match object; span=(29441, 29444), match='THF'>
<re.Match object; span=(29447, 29453), match='CH2THF'>
<re.Match object; span=(29486, 29492), match='CH3THF'>
<re.Match object; span=(29657, 29663), match='CH2THF'>
<re.Match object; span=(29882, 29885), match='UMP'>
<re.Match object; span=(29960, 29966), match='CH2THF'>
<re.Match object; span=(30064, 30067), match='TMP'>
<re.Match object; span=(30101, 30104), match='DNA'>
<re.Match object; span=(30420, 30425), match='MTHFR'>
<re.Match object; span=(30762, 30767), match='MTHFR'>
<re.Match object; span=(30913, 30918), match='MTHFR'>
<re.Match object; span=(31530, 31535), match='MTHFR'>
<re.Match object; span=(31652, 31657), match='MTHFR'>
<re.Match object; span=(31990, 31995), match='MTHFR'>
<re.Match object; span=(32029, 32035), match='FOLFOX'>
<re.Match object; span=(32061, 32066), match='MTHFR'>
<re.Match object; span=(32368, 32373), match='MTHFR'>
<re.Match object; span=(32548, 32553), match='MTHFR'>
<re.Match object; span=(32923, 32930), match='DPYD*2A'>
<re.Match object; span=(33023, 33027), match='DPYD'>
<re.Match object; span=(33377, 33382), match='MTHFR'>
<re.Match object; span=(34456, 34464), match='GLOBOCAN'>
<re.Match object; span=(34519, 34523), match='IARC'>
<re.Match object; span=(34883, 34886), match='III'>
<re.Match object; span=(35744, 35747), match='III'>
<re.Match object; span=(35769, 35776), match='FOLFIRI'>
<re.Match object; span=(35784, 35791), match='FOLFOX4'>
<re.Match object; span=(36357, 36360), match='CAO'>
<re.Match object; span=(36361, 36364), match='ARO'>
<re.Match object; span=(36365, 36368), match='AIO'>
<re.Match object; span=(36390, 36393), match='III'>
<re.Match object; span=(37276, 37279), match='BMC'>
<re.Match object; span=(37886, 37889), match='GKB'>
<re.Match object; span=(38264, 38269), match='IVS14'>
<re.Match object; span=(38769, 38776), match='FOLFOX6'>
<re.Match object; span=(39540, 39545), match='MTHFR'>
<re.Match object; span=(40008, 40011), match='WMA'>
<re.Match object; span=(40085, 40088), match='WMA'>
<re.Match object; span=(40403, 40408), match='RECST'>
<re.Match object; span=(40451, 40467), match='RECISTGuidelines'>
<re.Match object; span=(40588, 40594), match='CYP2A6'>
<re.Match object; span=(40600, 40606), match='CYP1A1'>
<re.Match object; span=(41042, 41048), match='FOLFOX'>
<re.Match object; span=(41521, 41524), match='ABP'>
<re.Match object; span=(42213, 42216), match='DPD'>
<re.Match object; span=(42254, 42257), match='DPD'>
<re.Match object; span=(42587, 42591), match='DPYD'>
<re.Match object; span=(42592, 42597), match='IVS14'>
<re.Match object; span=(43610, 43614), match='PLoS'>
<re.Match object; span=(43707, 43711), match='TYMS'>
<re.Match object; span=(43750, 43754), match='DPYD'>
<re.Match object; span=(43981, 43985), match='DPYD'>
<re.Match object; span=(44565, 44570), match='MTHFR'>
<re.Match object; span=(45502, 45507), match='MTHFR'>
<re.Match object; span=(45534, 45540), match='FOLFOX'>
Capecitabine:
- Extraction of variants associate to the known genes.
- Steps:
- Download the known clinical annotation and variant annotation from pharmgkb.
- Merge the two dataframe to ascertain the unique snps ids for the known genes.
- Scrape the allele frequency based on the known snps ids from ncbi database.
#### variant annotation all-data_variants_capecitabine.tsv
path_capecitabine = "/content/drive/MyDrive/Yemaachi_works/Capecitabine/"
file_clinical = "/content/drive/MyDrive/Yemaachi_works/Capecitabine/all-data_clinical_capecitabine.tsv"
file_variant = "/content/drive/MyDrive/Yemaachi_works/Capecitabine/all-data_variants_capecitabine.tsv"
### dataframe
data_variant = pd.read_csv(file_variant,sep="\t")
data_clinical = pd.read_csv(file_clinical,sep="\t")
### visualize the first three features:
data_variant.head(3)
|
PharmGKB ID |
Variant |
Literature |
Genes |
Association |
Significance |
P-Value |
# of Cases |
# of Controls |
Biogeographical Groups |
Phenotype Categories |
Pediatric |
More Details |
Molecules |
| 0 |
1449731693 |
rs3918290 |
PMID:30114658 |
DPYD |
Genotype CT is associated with increased Drug ... |
yes |
< 0.001 |
2105.0 |
NaN |
Unknown |
Toxicity |
False |
Grade >=2 lethargy, diarrhea, stomatitis and h... |
capecitabine; fluorouracil |
| 1 |
1448568402 |
rs1801158 |
PMID:27995989 |
DPYD |
Genotype CT is not associated with risk of Dru... |
no |
> 0.05 |
185.0 |
NaN |
Unknown |
Toxicity |
False |
No significant association with global toxicit... |
bevacizumab; capecitabine; cisplatin; docetaxe... |
| 2 |
827823452 |
rs3918290 |
PMID:19530960 |
DPYD |
Genotype CT is not associated with Drug Toxici... |
no |
= 1.0 |
111.0 |
NaN |
European |
Toxicity |
False |
Note: only one heterozygote for this variant w... |
capecitabine; fluorouracil |
<svg xmlns="http://www.w3.org/2000/svg" height="24px"viewBox="0 0 24 24"
width="24px">
<script>
const buttonEl =
document.querySelector('#df-604ca3d7-b0d7-40cd-bde8-5068fa241c68 button.colab-df-convert');
buttonEl.style.display =
google.colab.kernel.accessAllowed ? 'block' : 'none';
async function convertToInteractive(key) {
const element = document.querySelector('#df-604ca3d7-b0d7-40cd-bde8-5068fa241c68');
const dataTable =
await google.colab.kernel.invokeFunction('convertToInteractive',
[key], {});
if (!dataTable) return;
const docLinkHtml = 'Like what you see? Visit the ' +
'<a target="_blank" href=https://colab.research.google.com/notebooks/data_table.ipynb>data table notebook</a>'
+ ' to learn more about interactive tables.';
element.innerHTML = '';
dataTable['output_type'] = 'display_data';
await google.colab.output.renderOutput(dataTable, element);
const docLink = document.createElement('div');
docLink.innerHTML = docLinkHtml;
element.appendChild(docLink);
}
</script>
</div>
data_variant.shape
(486, 14)
### merge dataframe: variant and clinical annotations
#pd.merge(cyclo_clinical_data,cyclo_variants_data,on="Variant",how="outer")
#merge_data = pd.merge(data_clinical,data_variant,on=["Variant"],how="inner").drop_duplicates(["Variant"]).reset_index(drop=True)
data_variant.Genes.value_counts()
DPYD 239
MTHFR 27
TYMS 26
CDA 20
ERCC2 10
...
PTGS2 1
SLC28A1 1
PHC1 1
FAT1 1
UGT1A1 1
Name: Genes, Length: 78, dtype: int64
data_variant.columns
Index(['PharmGKB ID', 'Variant', 'Literature', 'Genes', 'Association',
'Significance', 'P-Value', '# of Cases', '# of Controls',
'Biogeographical Groups', 'Phenotype Categories', 'Pediatric',
'More Details', 'Molecules'],
dtype='object')
data_variants_capecitabine = data_variant[["PharmGKB ID","Variant","Literature","Genes","Phenotype Categories","Molecules"]]
data_variants_capecitabine.head()
|
PharmGKB ID |
Variant |
Literature |
Genes |
Phenotype Categories |
Molecules |
| 0 |
1449731693 |
rs3918290 |
PMID:30114658 |
DPYD |
Toxicity |
capecitabine; fluorouracil |
| 1 |
1448568402 |
rs1801158 |
PMID:27995989 |
DPYD |
Toxicity |
bevacizumab; capecitabine; cisplatin; docetaxe... |
| 2 |
827823452 |
rs3918290 |
PMID:19530960 |
DPYD |
Toxicity |
capecitabine; fluorouracil |
| 3 |
1448568409 |
rs2612091 |
PMID:27995989 |
ENOSF1 |
Toxicity |
bevacizumab; capecitabine; cisplatin; docetaxe... |
| 4 |
827817287 |
rs9937 |
PMID:22026922 |
RRM1 |
Efficacy |
capecitabine; cisplatin; docetaxel; epirubicin... |
<svg xmlns="http://www.w3.org/2000/svg" height="24px"viewBox="0 0 24 24"
width="24px">
<script>
const buttonEl =
document.querySelector('#df-a2826277-d7eb-4bc2-9d6f-efbd85dce33e button.colab-df-convert');
buttonEl.style.display =
google.colab.kernel.accessAllowed ? 'block' : 'none';
async function convertToInteractive(key) {
const element = document.querySelector('#df-a2826277-d7eb-4bc2-9d6f-efbd85dce33e');
const dataTable =
await google.colab.kernel.invokeFunction('convertToInteractive',
[key], {});
if (!dataTable) return;
const docLinkHtml = 'Like what you see? Visit the ' +
'<a target="_blank" href=https://colab.research.google.com/notebooks/data_table.ipynb>data table notebook</a>'
+ ' to learn more about interactive tables.';
element.innerHTML = '';
dataTable['output_type'] = 'display_data';
await google.colab.output.renderOutput(dataTable, element);
const docLink = document.createElement('div');
docLink.innerHTML = docLinkHtml;
element.appendChild(docLink);
}
</script>
</div>
data_variants_capecitabine.Genes.value_counts()
DPYD 239
MTHFR 27
TYMS 26
CDA 20
ERCC2 10
...
PTGS2 1
SLC28A1 1
PHC1 1
FAT1 1
UGT1A1 1
Name: Genes, Length: 78, dtype: int64
data_snps_capecitabine = data_variants_capecitabine[data_variants_capecitabine.Variant.str.startswith("rs")]
data_snps_capecitabine.shape
(472, 6)
data_snps_capecitabine.to_csv("/content/drive/MyDrive/Yemaachi_works/Capecitabine/variants_gene_pairs.csv",index = False)
### save the dataframe:
#new_data_variant.to_csv("/content/drive/MyDrive/Yemaachi_works/Capecitabine/all_new__capecitabine_updated.csv",index=False)
###https://www.ncbi.nlm.nih.gov/snp/rs11615/download/frequency
"https://docs.google.com/spreadsheets/d/1kxbmDslwLEzvPFQ0n_c7JAOt6u1MU4zl9brHvL5ea98/edit#gid=1101517327"
Access the frequency Table:
%cd "Cyclophosphamide "
/content/drive/MyDrive/Yemaachi_works/Cyclophosphamide
import gspread
import pandas as pd
from google.colab import auth
from oauth2client.client import GoogleCredentials
auth.authenticate_user()
gc = gspread.authorize(GoogleCredentials.get_application_default())
worksheet = gc.open('variants_genes_pairs_cyclophosphamide').sheet1
rows = worksheet.get_all_values()
df_variant_genes_cyclophosphamide = pd.DataFrame.from_records(rows[1:],columns=rows[0])
df_variant_genes_cyclophosphamide.head(10)
|
PharmGKB ID |
Variant |
Literature |
Genes |
Phenotype Categories |
Molecules |
Gene_consequence |
| 0 |
769245657 |
rs1045642 |
PMID:20638924 |
ABCB1 |
Toxicity |
cyclophosphamide; fluorouracil |
ABCB1 : Missense Variant |
| 1 |
769245656 |
rs45445694 |
PMID:20638924 |
TYMS |
Toxicity |
cyclophosphamide; fluorouracil |
TYMSOS : Intron Variant TYMS : 5 Prime UTR Var... |
| 2 |
699642290 |
rs4880 |
PMCID:PMC2697269 |
SOD2 |
Efficacy |
cyclophosphamide |
SOD2 : Missense Variant |
| 3 |
1184233620 |
rs9611280 |
PMCID:PMC3948785 |
TNRC6B |
Toxicity |
asparaginase; cyclophosphamide; cytarabine; da... |
TNRC6B : Missense Variant |
| 4 |
1446904891 |
rs3829306 |
PMID:24599932 |
SLCO1B1 |
Toxicity |
cyclophosphamide; epirubicin; paclitaxel |
SLCO1B1 : Intron Variant |
| 5 |
827828575 |
rs4244285 |
PMID:20358205 |
CYP2C19 |
Toxicity |
cyclophosphamide |
CYP2C19 : Synonymous Variant |
| 6 |
769245648 |
rs1801133 |
PMID:20638924 |
MTHFR |
Toxicity |
cyclophosphamide; fluorouracil |
MTHFR : Missense Variant |
| 7 |
1184233630 |
rs197388 |
PMCID:PMC3948785 |
DDX20 |
Toxicity |
asparaginase; cyclophosphamide; cytarabine; da... |
INKA2 : Intron Variant DDX20 : 2KB Upstream Va... |
| 8 |
769245653 |
rs1042522 |
PMID:20638924 |
TP53 |
Toxicity |
cyclophosphamide; fluorouracil |
TP53 : Missense Variant |
| 9 |
1446904882 |
rs6473187 |
PMID:24599932 |
SPIDR |
Toxicity |
cyclophosphamide; epirubicin; paclitaxel |
SPIDR : Intron Variant |
<svg xmlns="http://www.w3.org/2000/svg" height="24px"viewBox="0 0 24 24"
width="24px">
<script>
const buttonEl =
document.querySelector('#df-b5ca3269-808b-4d58-b4c9-1f2934ea177a button.colab-df-convert');
buttonEl.style.display =
google.colab.kernel.accessAllowed ? 'block' : 'none';
async function convertToInteractive(key) {
const element = document.querySelector('#df-b5ca3269-808b-4d58-b4c9-1f2934ea177a');
const dataTable =
await google.colab.kernel.invokeFunction('convertToInteractive',
[key], {});
if (!dataTable) return;
const docLinkHtml = 'Like what you see? Visit the ' +
'<a target="_blank" href=https://colab.research.google.com/notebooks/data_table.ipynb>data table notebook</a>'
+ ' to learn more about interactive tables.';
element.innerHTML = '';
dataTable['output_type'] = 'display_data';
await google.colab.output.renderOutput(dataTable, element);
const docLink = document.createElement('div');
docLink.innerHTML = docLinkHtml;
element.appendChild(docLink);
}
</script>
</div>
df_variant_genes_cyclophosphamide.Genes.value_counts()
PIK3R2 13
CYP2B6 8
ABCC1 8
ABCB1 7
VEGFA 6
..
MTR 1
MIR1307 1
TP53AIP1 1
MIR618 1
CYP4X1 1
Name: Genes, Length: 117, dtype: int64
df_variant_genes_cyclophosphamide[df_variant_genes_cyclophosphamide.Genes== ""]
|
PharmGKB ID |
Variant |
Literature |
Genes |
Phenotype Categories |
Molecules |
Gene_consequence |
| 25 |
1184233654 |
rs56103835 |
PMCID:PMC3948785 |
|
Toxicity |
asparaginase; cyclophosphamide; cytarabine; da... |
MIR323B : Non Coding Transcript Variant |
| 70 |
1447676969 |
rs80223967 |
PMCID:PMC4742546 |
|
Toxicity |
cyclophosphamide; cytarabine; daunorubicin; de... |
LOC105372912 : Intron Variant |
| 85 |
1447676983 |
rs17021408 |
PMCID:PMC4742546 |
|
Toxicity |
cyclophosphamide; cytarabine; daunorubicin; de... |
LOC105372912 : Intron Variant |
| 114 |
1448624269 |
rs11636687 |
PMCID:PMC5652844 |
|
Toxicity |
cyclophosphamide; epirubicin; fluorouracil |
None |
| 152 |
1448624409 |
rs4896870 |
PMCID:PMC5652844 |
|
Toxicity |
cyclophosphamide; epirubicin; fluorouracil |
None |
| 183 |
1447676926 |
rs1891059 |
PMCID:PMC4742546 |
|
Toxicity |
cyclophosphamide; cytarabine; daunorubicin; de... |
LOC105372912 : Intron Variant |
<svg xmlns="http://www.w3.org/2000/svg" height="24px"viewBox="0 0 24 24"
width="24px">
<script>
const buttonEl =
document.querySelector('#df-bf79503e-9f65-480f-b6b2-3dedad02a868 button.colab-df-convert');
buttonEl.style.display =
google.colab.kernel.accessAllowed ? 'block' : 'none';
async function convertToInteractive(key) {
const element = document.querySelector('#df-bf79503e-9f65-480f-b6b2-3dedad02a868');
const dataTable =
await google.colab.kernel.invokeFunction('convertToInteractive',
[key], {});
if (!dataTable) return;
const docLinkHtml = 'Like what you see? Visit the ' +
'<a target="_blank" href=https://colab.research.google.com/notebooks/data_table.ipynb>data table notebook</a>'
+ ' to learn more about interactive tables.';
element.innerHTML = '';
dataTable['output_type'] = 'display_data';
await google.colab.output.renderOutput(dataTable, element);
const docLink = document.createElement('div');
docLink.innerHTML = docLinkHtml;
element.appendChild(docLink);
}
</script>
</div>
known_genes_cyclophosphamide = df_variant_genes_cyclophosphamide[df_variant_genes_cyclophosphamide.Genes != ""]
known_genes_cyclophosphamide.shape
(195, 7)
#### single drug: cyclophosphamide
known_genes_cyclophosphamide[known_genes_cyclophosphamide["Molecules"] == "cyclophosphamide"].shape[0]
20
#### combine drugs:
known_genes_cyclophosphamide[known_genes_cyclophosphamide["Molecules"] != "cyclophosphamide"].shape[0]
175
from collections import Counter
sorted(Counter(known_genes_cyclophosphamide.Genes).items(),key= lambda x: x[1] ,reverse=True)
[('PIK3R2', 13),
('CYP2B6', 8),
('ABCC1', 8),
('ABCB1', 7),
('VEGFA', 6),
('ALDH1A1', 5),
('DROSHA', 4),
('ABCC2', 4),
('PERP', 4),
('SLC22A16', 4),
('CYP2C19', 3),
('XRCC1', 3),
('GSTM3', 3),
('PIGB', 3),
('EPAS1', 3),
('HMMR', 3),
('SLCO1B1', 2),
('MTHFR', 2),
('TP53', 2),
('GSTP1', 2),
('NQO1', 2),
('GSTA1', 2),
('CBR3', 2),
('CYP2E1', 2),
('ERCC2', 2),
('NOS3', 2),
('ERCC1', 2),
('RAB27A', 2),
('CTH', 2),
('INSR', 2),
('TYMS', 1),
('SOD2', 1),
('TNRC6B', 1),
('DDX20', 1),
('SPIDR', 1),
('CNOT1', 1),
('CYP3A4', 1),
('CYP1B1', 1),
('TNRC6A', 1),
('IKZF3', 1),
('XPO5', 1),
('GATA3', 1),
('MIR449B', 1),
('UGT2B7', 1),
('MIR423', 1),
('MCPH1', 1),
('MIR2053', 1),
('CYBA', 1),
('GEMIN4', 1),
('RBX1', 1),
('MIR300', 1),
('AGO1', 1),
('MIR577', 1),
('ADH1C', 1),
('CXCL8', 1),
('MIR1206', 1),
('LINC00251', 1),
('MIR492', 1),
('MIR604', 1),
('CYP2C8', 1),
('CXCR2', 1),
('PNPLA3', 1),
('NCF4', 1),
('MIR618', 1),
('TP53AIP1', 1),
('MIR1307', 1),
('MTR', 1),
('MIR146A', 1),
('GNL3', 1),
('TOP2A', 1),
('MISP', 1),
('DOK5', 1),
('NOS1', 1),
('EPHA6', 1),
('EPHX1', 1),
('NAT2', 1),
('EGLN3', 1),
('RAD52', 1),
('MUTYH', 1),
('ABCG2', 1),
('MIR4268', 1),
('ABCC4', 1),
('CYP3A5', 1),
('ALDH3A1', 1),
('FLT1', 1),
('UGT1A1', 1),
('CYP2E1; DUX1', 1),
('C5orf22', 1),
('TNFSF13B', 1),
('ABCC3', 1),
('CTNNB1', 1),
('TUBB2A', 1),
('ATM', 1),
('FCGR3A', 1),
('FOXO1', 1),
('FGFR4', 1),
('RAC2', 1),
('DPYD', 1),
('ERCC5', 1),
('RRM2B', 1),
('MIR3117', 1),
('LIG3', 1),
('ATF5', 1),
('FCGR2A', 1),
('ZNF215', 1),
('NR1I2', 1),
('CXCL12', 1),
('PPP2R5D', 1),
('TPMT', 1),
('CCNK', 1),
('NQO2', 1),
('CBR1', 1),
('IRS1', 1),
('SLC28A3', 1),
('BMP7', 1),
('CYP4X1', 1)]
genes = []
frequency = []
from collections import Counter
for gene , freq in sorted(Counter(known_genes_cyclophosphamide.Genes).items(),key= lambda x: x[1] ,reverse=True)[:11]:
genes.append(gene)
frequency.append(freq)
from matplotlib import pyplot as plt
with plt.style.context("ggplot"):
plt.figure(figsize=(18,7))
plt.bar(genes,frequency,color="blue")
plt.xticks(rotation = 90)
plt.title("Frequency of Variants: Cyclophosphamide",fontsize=18)
plt.xlabel("Genes",fontsize=18)
plt.ylabel("Frequency",fontsize = 18)
plt.xticks(rotation = 45)
for i in range(len(frequency)):
plt.annotate(str(frequency[i]), xy=(genes[i],frequency[i]), ha='center', va='bottom')
plt.show()
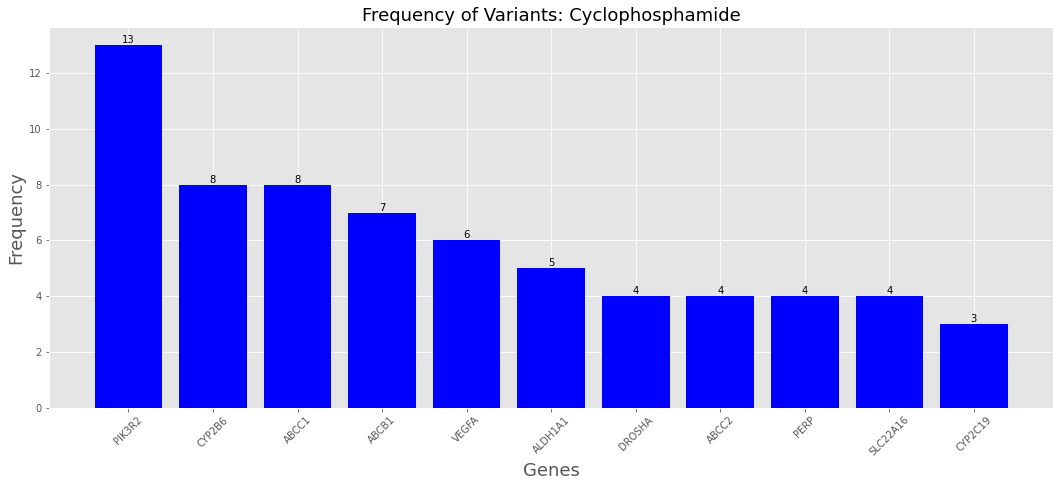
known_genes_cyclophosphamide["Phenotype Categories"].value_counts()
Toxicity 141
Efficacy 42
Metabolism/PK,Toxicity 4
Efficacy,Metabolism/PK,Toxicity 4
Metabolism/PK 4
Name: Phenotype Categories, dtype: int64
#### import tqdm library
import tqdm
from tqdm.notebook import tqdm
#### define a function that would run the process:
def frequency_table(*,snp_id = None,gene_name = None,gene_consequence = None):
### create access link to file
"""
Args:
snp_id: pass the snp_id for given gene
gene_name : pass the corresponding gene
gene_consequence: effect of that particular gene.
"""
first_link = "https://www.ncbi.nlm.nih.gov/snp/"
snp_id = str(snp_id)
last_link = "/download/frequency"
actual_link = first_link + snp_id + last_link ###### combine the names
try:
#### check for access of the url:
url_access = requests.get(actual_link)
text = url_access.text
text_list = text.split("#################")[1] ### split the raw text into two separate strings.
texts = text_list.split("\n")[1:-1]
data = pd.DataFrame([x.split("\t") for x in texts]) ### convert the raw text into dataframe
data.columns = data.iloc[0] ### set the first row as the header
data = data.drop(data.index[0]) ### drop the row with the header:
data["gene"] = gene_name
data["gene_consequence"] = gene_consequence
data["snp_id"] = snp_id
return data
except:
print(f"There is no info for the snp_id entered: {snp_id}")
data = []
return data
#### compile all the data
def compile_data(snps_data = None):
"""
Takes the compile data:
"""
list_of_data = []
for index, rows in tqdm(snps_data.iterrows()):
data = frequency_table(snp_id = rows["Variant"],gene_name = rows["Genes"], gene_consequence = rows["Gene_consequence"])
if type(data) == pd.core.frame.DataFrame:
data = data
else:
continue
list_of_data.append(data)
data_allele_frequency = pd.concat(list_of_data)
"""
Return:
the compiled dataset
"""
return data_allele_frequency
#### run the dataframe:
data = compile_data(snps_data = known_genes_cyclophosphamide)
0it [00:00, ?it/s]
There is no info for the snp_id entered: rs1799735
### reset the data by drop unordered index.
data = data.reset_index(drop=True)
data_cyclophosphamide = data
data_cyclophosphamide.head()
|
#Study |
Population |
Group |
Samplesize |
Ref Allele |
Alt Allele |
BioProject ID |
BioSample ID |
gene |
gene_consequence |
snp_id |
| 0 |
TopMed |
Global |
Study-wide |
264690 |
A=0.400892 |
G=0.599108 |
PRJNA400167 |
|
ABCB1 |
ABCB1 : Missense Variant |
rs1045642 |
| 1 |
gnomAD - Exomes |
Global |
Study-wide |
251312 |
A=0.495703 |
G=0.504297 |
PRJNA398795 |
SAMN07488253 |
ABCB1 |
ABCB1 : Missense Variant |
rs1045642 |
| 2 |
gnomAD - Exomes |
European |
Sub |
135256 |
A=0.546201 |
G=0.453799 |
|
SAMN10181265 |
ABCB1 |
ABCB1 : Missense Variant |
rs1045642 |
| 3 |
gnomAD - Exomes |
Asian |
Sub |
48998 |
A=0.51590 |
G=0.48410 |
|
|
ABCB1 |
ABCB1 : Missense Variant |
rs1045642 |
| 4 |
gnomAD - Exomes |
American |
Sub |
34588 |
A=0.45186 |
G=0.54814 |
|
SAMN07488255 |
ABCB1 |
ABCB1 : Missense Variant |
rs1045642 |
<svg xmlns="http://www.w3.org/2000/svg" height="24px"viewBox="0 0 24 24"
width="24px">
<script>
const buttonEl =
document.querySelector('#df-bf14b26f-0ded-49e9-9100-ce37e6d64d0a button.colab-df-convert');
buttonEl.style.display =
google.colab.kernel.accessAllowed ? 'block' : 'none';
async function convertToInteractive(key) {
const element = document.querySelector('#df-bf14b26f-0ded-49e9-9100-ce37e6d64d0a');
const dataTable =
await google.colab.kernel.invokeFunction('convertToInteractive',
[key], {});
if (!dataTable) return;
const docLinkHtml = 'Like what you see? Visit the ' +
'<a target="_blank" href=https://colab.research.google.com/notebooks/data_table.ipynb>data table notebook</a>'
+ ' to learn more about interactive tables.';
element.innerHTML = '';
dataTable['output_type'] = 'display_data';
await google.colab.output.renderOutput(dataTable, element);
const docLink = document.createElement('div');
docLink.innerHTML = docLinkHtml;
element.appendChild(docLink);
}
</script>
</div>
data_cyclophosphamide.to_csv("cyclophosphamide_alle_frequency.csv",index = False)
%pwd
'/content/drive/MyDrive/Yemaachi_works/Cyclophosphamide '
%cd ..
%pwd
/content/drive/MyDrive/Yemaachi_works
'/content/drive/MyDrive/Yemaachi_works'
Analyse the known genes for capecitabine:
- Download both clinical and variants data from pharmgkb database.
worksheet = gc.open('variants_gene_pairs').sheet1
rows = worksheet.get_all_values()
df_variant_genes_capecitabine = pd.DataFrame.from_records(rows[1:],columns=rows[0])
df_variant_genes_capecitabine.head(10)
|
PharmGKB ID |
Variant |
Literature |
Genes |
Phenotype Categories |
Molecules |
Gene_consequence |
| 0 |
1449731693 |
rs3918290 |
PMID:30114658 |
DPYD |
Toxicity |
capecitabine; fluorouracil |
DPYD : Splice Donor Variant |
| 1 |
1448568402 |
rs1801158 |
PMID:27995989 |
DPYD |
Toxicity |
bevacizumab; capecitabine; cisplatin; docetaxe... |
DPYD : Missense Variant |
| 2 |
1448568409 |
rs2612091 |
PMID:27995989 |
ENOSF1 |
Toxicity |
bevacizumab; capecitabine; cisplatin; docetaxe... |
ENOSF1 : Intron Variant |
| 3 |
827817287 |
rs9937 |
PMID:22026922 |
RRM1 |
Efficacy |
capecitabine; cisplatin; docetaxel; epirubicin... |
RRM1 : Synonymous Variant |
| 4 |
1448568388 |
rs11479 |
PMID:27995989 |
TYMP |
Toxicity |
bevacizumab; capecitabine; cisplatin; docetaxe... |
TYMP : Stop Gained SCO2 : Intron Variant |
| 5 |
1451147880 |
rs1801159 |
PMID:32378051 |
DPYD |
Toxicity |
capecitabine |
DPYD : Missense Variant |
| 6 |
1448568395 |
rs56038477 |
PMID:27995989 |
DPYD |
Toxicity |
bevacizumab; capecitabine; cisplatin; docetaxe... |
DPYD : Synonymous Variant |
| 7 |
1446908428 |
rs61764370 |
PMCID:PMC4551162 |
KRAS |
Efficacy |
capecitabine; cetuximab; oxaliplatin |
KRAS : 3 Prime UTR Variant |
| 8 |
1184511648 |
rs45445694 |
PMID:23263912 |
TYMS |
Efficacy |
capecitabine; paclitaxel |
TYMSOS : Intron Variant TYMS : 5 Prime UTR Var... |
| 9 |
1450953284 |
rs1801265 |
PMID:20819423 |
DPYD |
Toxicity |
capecitabine; fluorouracil |
DPYD : Missense Variant |
<svg xmlns="http://www.w3.org/2000/svg" height="24px"viewBox="0 0 24 24"
width="24px">
<script>
const buttonEl =
document.querySelector('#df-361df778-4a58-4546-85d6-96a5ee0250e0 button.colab-df-convert');
buttonEl.style.display =
google.colab.kernel.accessAllowed ? 'block' : 'none';
async function convertToInteractive(key) {
const element = document.querySelector('#df-361df778-4a58-4546-85d6-96a5ee0250e0');
const dataTable =
await google.colab.kernel.invokeFunction('convertToInteractive',
[key], {});
if (!dataTable) return;
const docLinkHtml = 'Like what you see? Visit the ' +
'<a target="_blank" href=https://colab.research.google.com/notebooks/data_table.ipynb>data table notebook</a>'
+ ' to learn more about interactive tables.';
element.innerHTML = '';
dataTable['output_type'] = 'display_data';
await google.colab.output.renderOutput(dataTable, element);
const docLink = document.createElement('div');
docLink.innerHTML = docLinkHtml;
element.appendChild(docLink);
}
</script>
</div>
df_variant_genes_capecitabine.Genes.value_counts()
DPYD 40
SPARC 8
HLA-G 8
CDA 7
UMPS 6
..
PTGS2 1
ADCY2 1
XRCC1 1
SLC28A1 1
CYP1A1 1
Name: Genes, Length: 72, dtype: int64
df_variant_genes_capecitabine.Genes.unique()
array(['DPYD', 'ENOSF1', 'RRM1', 'TYMP', 'KRAS', 'TYMS', 'CYP1B1',
'SMAD7', 'CDA', 'GSTP1', 'ABCB1', 'MTHFR', 'VEGFA', 'CES2',
'MIR27A', 'ERCC1', 'HLA-G', 'MGAT4A', 'SLCO1B1', 'ERCC2',
'SLC19A1', 'UMPS', 'REV3L', 'MTR', 'SHMT1', 'SLC22A7', 'TK1',
'EXO1', 'PTEN', 'CYP19A1', 'SELE', 'UPB1', 'MTHFD1', 'WDR7',
'DLG5', 'ENOSF1; TYMS', 'NSUN3', 'CDX2', 'PTGS2', 'ADCY2', 'XRCC1',
'SLC28A1', 'PHC1', 'ABCC11', 'FAT1', 'TENM4', 'ANK3', 'DPYS',
'MTRR', 'SMARCAD1', 'SSU72', 'MAN1A1', 'AREG', 'SPRY2', 'VPS13D',
'TP53', 'NCOA7', 'MIR2054', 'CD96', 'ZMIZ1', 'CCDC77', 'ADGRG7',
'CES1', 'CES1P1', 'SPARC', 'TMEM131L', 'SIRPA', 'LMNTD1', 'CCDC70',
'APOBEC2', 'ARHGEF4', 'CYP1A1'], dtype=object)
cape_genes = []
cape_frequency = []
for gene , freq in sorted(Counter(df_variant_genes_capecitabine.Genes).items(),key= lambda x: x[1] ,reverse=True)[:11]:
cape_genes.append(gene)
cape_frequency.append(freq)
with plt.style.context("ggplot"):
plt.figure(figsize=(12,7))
plt.bar(cape_genes,cape_frequency,color="blue")
plt.xticks(rotation = 90)
plt.title("Frequency of Variants: capecitabine",fontsize=18)
plt.xlabel("Genes",fontsize=18)
plt.ylabel("Frequency",fontsize = 18)
plt.xticks(rotation = 45)
for i in range(len(cape_frequency)):
plt.annotate(str(cape_frequency[i]), xy=(cape_genes[i],cape_frequency[i]), ha='center', va='bottom')
plt.show()
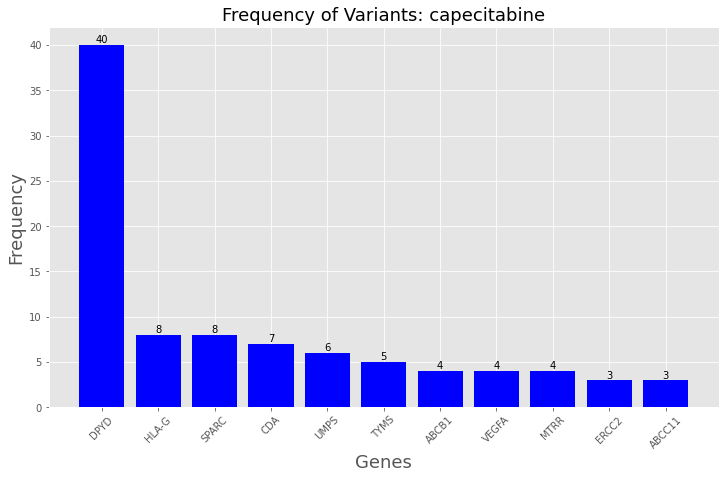
df_variant_genes_capecitabine["Phenotype Categories"].value_counts()
Toxicity 103
Efficacy 63
Metabolism/PK 2
Dosage,Toxicity 1
Name: Phenotype Categories, dtype: int64
df_variant_genes_capecitabine[df_variant_genes_capecitabine["Molecules"] == "capecitabine"].shape
(76, 7)
df_variant_genes_capecitabine[df_variant_genes_capecitabine["Molecules"] != "capecitabine"].shape
(93, 7)
data_capecitabine = compile_data(snps_data = df_variant_genes_capecitabine)
0it [00:00, ?it/s]
data_capecitabine.head()
|
#Study |
Population |
Group |
Samplesize |
Ref Allele |
Alt Allele |
BioProject ID |
BioSample ID |
gene |
gene_consequence |
snp_id |
| 1 |
gnomAD - Genomes |
Global |
Study-wide |
140212 |
C=0.995257 |
T=0.004743 |
PRJNA398795 |
SAMN07488253 |
DPYD |
DPYD : Splice Donor Variant |
rs3918290 |
| 2 |
gnomAD - Genomes |
European |
Sub |
75944 |
C=0.99227 |
T=0.00773 |
|
SAMN10181265 |
DPYD |
DPYD : Splice Donor Variant |
rs3918290 |
| 3 |
gnomAD - Genomes |
African |
Sub |
42020 |
C=0.99941 |
T=0.00059 |
|
SAMN07488254 |
DPYD |
DPYD : Splice Donor Variant |
rs3918290 |
| 4 |
gnomAD - Genomes |
American |
Sub |
13640 |
C=0.99817 |
T=0.00183 |
|
SAMN07488255 |
DPYD |
DPYD : Splice Donor Variant |
rs3918290 |
| 5 |
gnomAD - Genomes |
Ashkenazi Jewish |
Sub |
3324 |
C=0.9937 |
T=0.0063 |
|
SAMN07488252 |
DPYD |
DPYD : Splice Donor Variant |
rs3918290 |
<svg xmlns="http://www.w3.org/2000/svg" height="24px"viewBox="0 0 24 24"
width="24px">
<script>
const buttonEl =
document.querySelector('#df-76ab8478-f7ed-4af8-b262-8b5e30363dd1 button.colab-df-convert');
buttonEl.style.display =
google.colab.kernel.accessAllowed ? 'block' : 'none';
async function convertToInteractive(key) {
const element = document.querySelector('#df-76ab8478-f7ed-4af8-b262-8b5e30363dd1');
const dataTable =
await google.colab.kernel.invokeFunction('convertToInteractive',
[key], {});
if (!dataTable) return;
const docLinkHtml = 'Like what you see? Visit the ' +
'<a target="_blank" href=https://colab.research.google.com/notebooks/data_table.ipynb>data table notebook</a>'
+ ' to learn more about interactive tables.';
element.innerHTML = '';
dataTable['output_type'] = 'display_data';
await google.colab.output.renderOutput(dataTable, element);
const docLink = document.createElement('div');
docLink.innerHTML = docLinkHtml;
element.appendChild(docLink);
}
</script>
</div>
data_capecitabine.to_csv("/content/drive/MyDrive/Yemaachi_works/Capecitabine/capecitabine_allele_frequency.csv",index = False)

