Semi-supervised Cancer Prognosis Classifier with Bayesian Variational Autoencoder (SCAN)
Summary
This repository provides implementations of SCAN as described in our paper: Learning from Small Medical Data - Robust Semi-supervised Cancer Prognosis Classifier with Bayesian Variational Autoencoder. SCAN incorporates several deep learning approaches, including semi-supervised learning, ensemble learning, and variational autoencoder, to efficiently utilize precious patient data. Various patients (labeled/unlabeled/missing clinical data) with different data modalities (microarray/clinical data) can well fit into this unified framework to jointly train a powerful classifier for cancer prognosis.
Flowchart
The shcematic below shows an overview of the paper. Breast and non-small cell lung cancer data cohorts were collected and preprocessed according to the same principles outlined in the corresponding papers1,2.
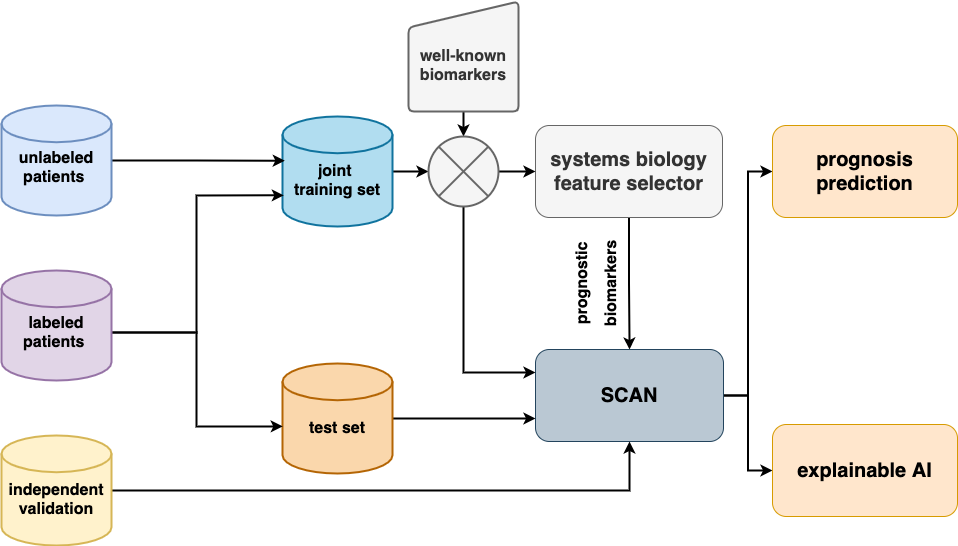
Key features of SCAN
In our previous research, we showed several deep learning approaches could potentially resolve commonly encountered issues when applied to biological applications such as curse of dimensionality1,2,3, censorship (missing labels)4, data scarcity5, and model robustness6.
Semi-supervised ensemble predictions
General framework to extract meaningful information from labeled/unlabeled multimodal data
Scalable light model ready for federated learning
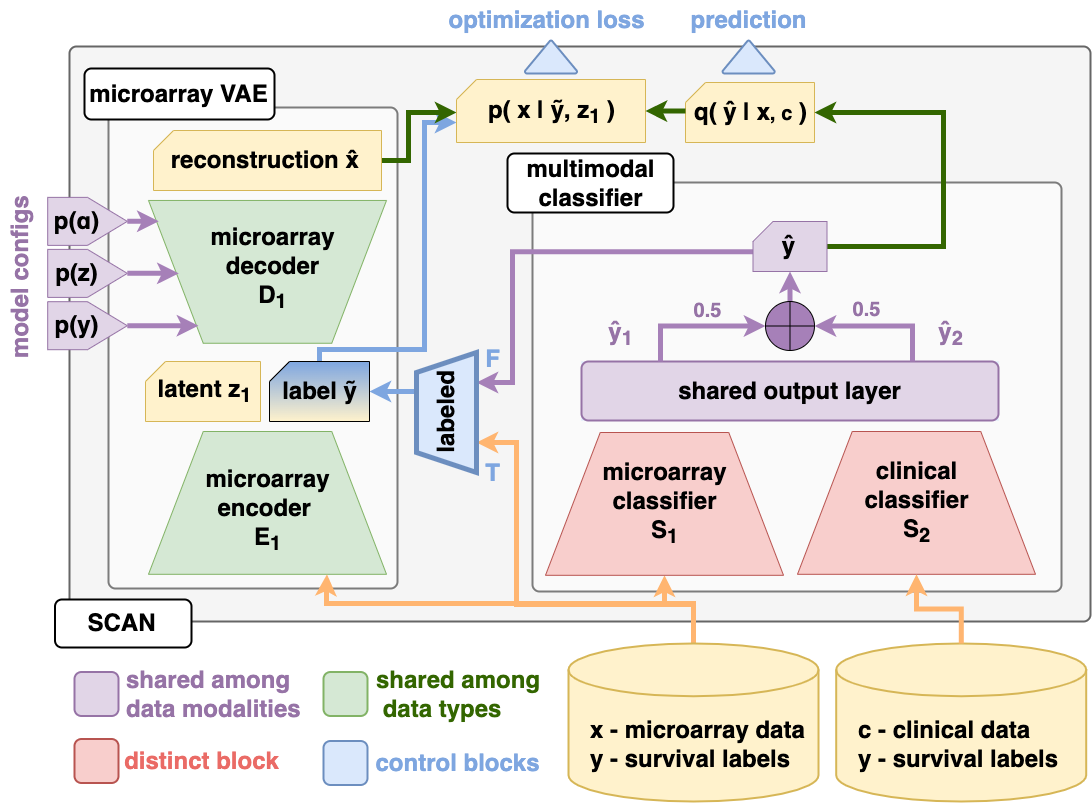
SCAN model architecture
Combining the design concepts mentioned above, we designed SCAN as shown in the figure below. For more details, please refer to our paper.
Basic usage
How to train SCAN
You can access SCAN models under various settings in /src:
python3 train_scan_breast.py # breast cancer
python3 train_scan_nsclc.py # non-small cell lung
The ensemble version of SCAN can be trained with:
bash train_scan_ens.sh
The trained models can be found in the corresponding directories in /model.
How to test/predict with SCAN
With trained models saved in /model, you can make predictions with it by:
python3 test_scan_breast.py
python3 test_scan_nsclc.py
python3 test_scan_ens.py # ensemble
Various performance evaluation illustration approaches
All figures & tables can be generated with /utils/plot_properties.py. Examples of performance illustrations can be found in /results.
Results
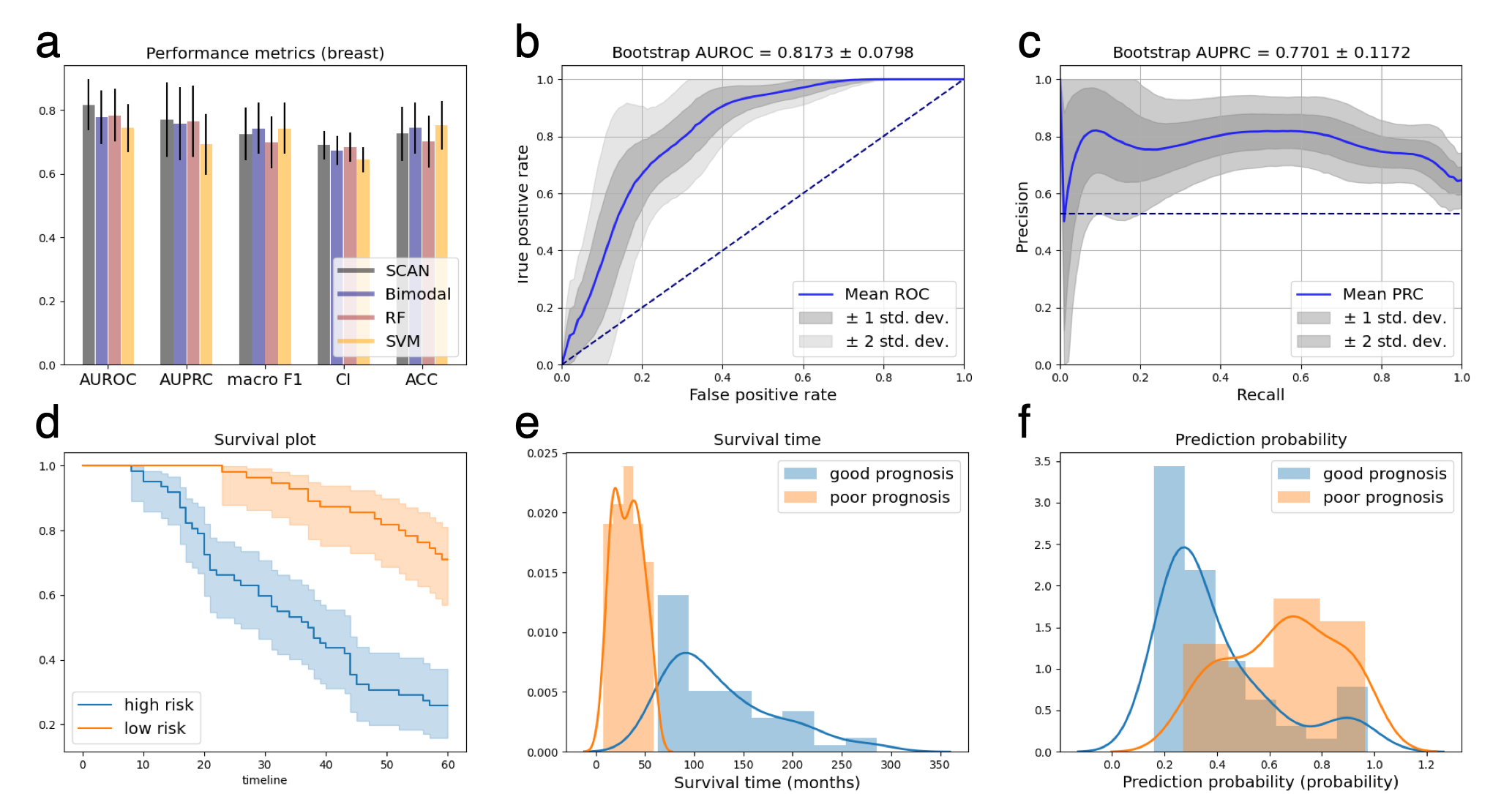
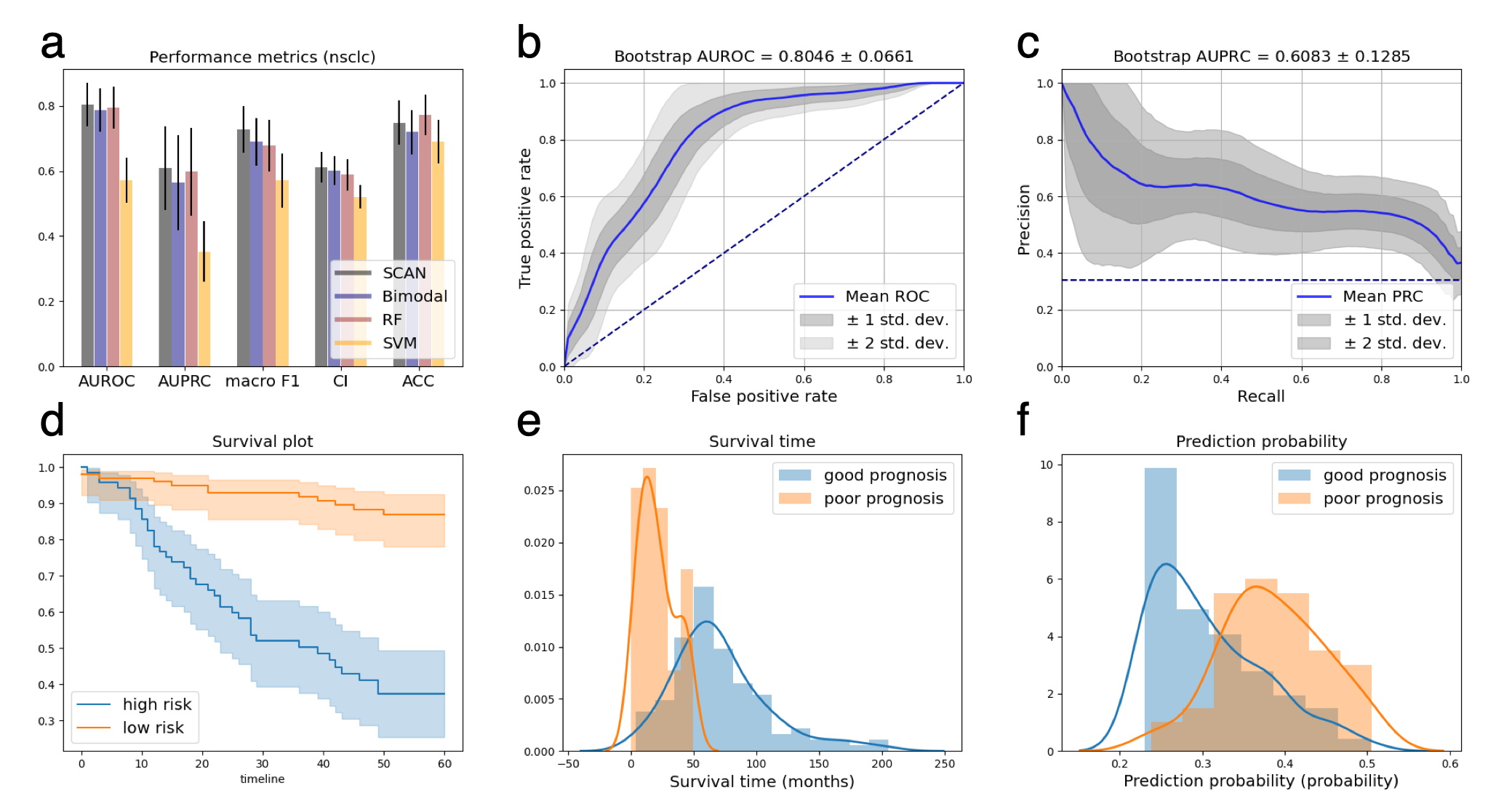
Experiment results showed that SCAN achieved better performance than various benchmarks.
Breast cancer perfomance summary
NSCLC cancer perfomance summary
Citation
Please cite our work if you find it useful for your research. Please feel free to contact us.
Reminder
For the trained models and breast cancer cohort (METABRIC), please download them from here.
References
[1] Cheng, L. H., Hsu, T. C., & Lin, C. (2021). Integrating ensemble systems biology feature selection and bimodal deep neural network for breast cancer prognosis prediction. Scientific Reports, 11(1), 1-10.
[2] Lai, Y. H., Chen, W. N., Hsu, T. C., Lin, C., Tsao, Y., & Wu, S. (2020). Overall survival prediction of non-small cell lung cancer by integrating microarray and clinical data with deep learning. Scientific reports, 10(1), 1-11.
[3] Liu, F. Y., Hsu, T. C., Choong, P., Lin, M. H., Chuang, Y. J., Chen, B. S., & Lin, C. (2018). Uncovering the regeneration strategies of zebrafish organs: a comprehensive systems biology study on heart, cerebellum, fin, and retina regeneration. BMC systems biology, 12(2), 33-46.
[4] Kingma, D. P., Mohamed, S., Jimenez Rezende, D., & Welling, M. (2014). Semi-supervised learning with deep generative models. Advances in neural information processing systems, 27.
[5] Hsu, T. C., & Lin, C. (2020, July). Generative adversarial networks for robust breast cancer prognosis prediction with limited data size. In 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) (pp. 5669-5672). IEEE.
[6] Hsu, T. C., & Lin, C. (2021, November). Training with Small Medical Data: Robust Bayesian Neural Networks for Colon Cancer Overall Survival Prediction. In 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) (pp. 2030-2033). IEEE.